visualization methods in ChIPseeker
After two weeks developed, I have added/updated some plot functions in ChIPseeker (version >=1.0.1).
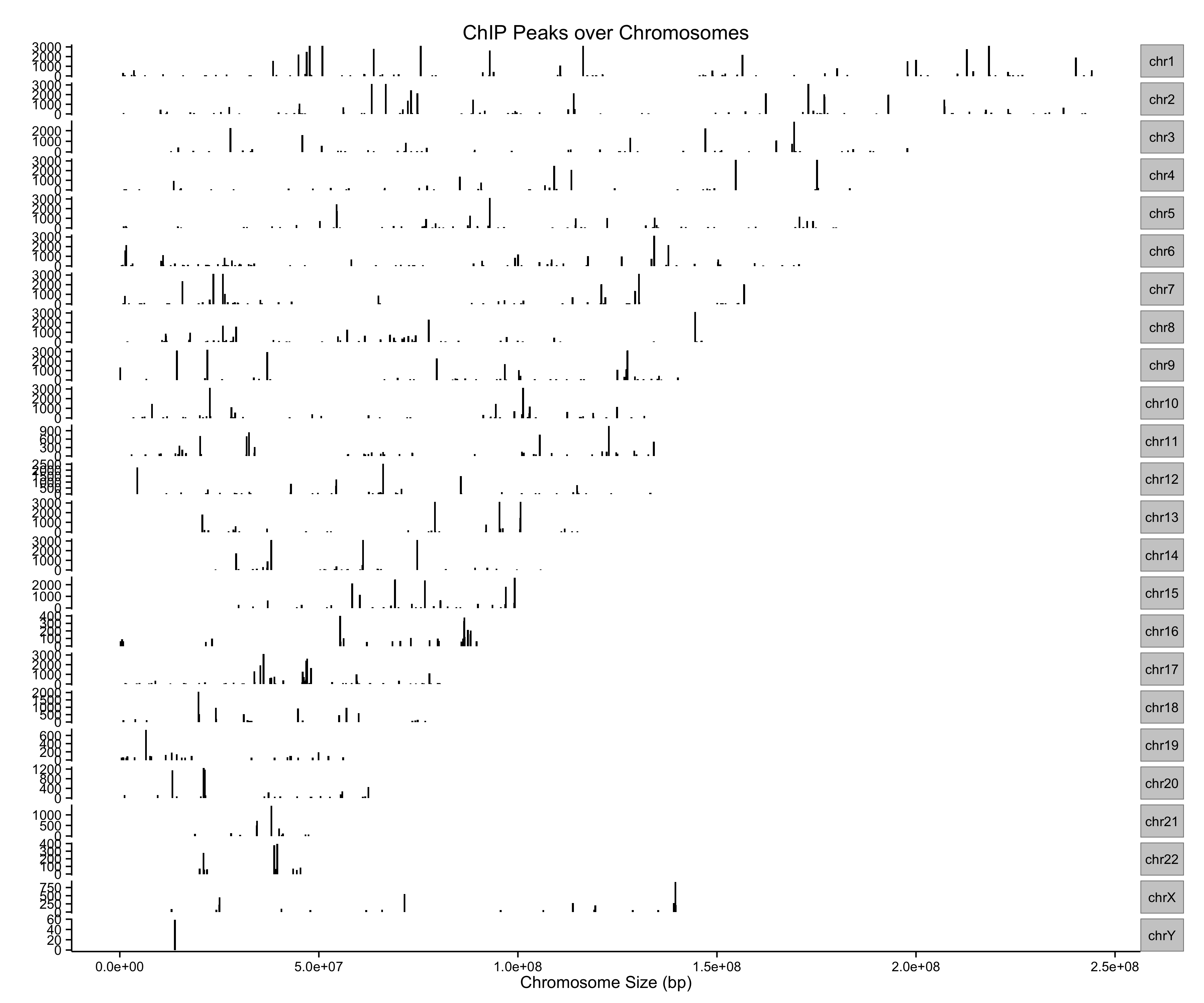
ChIP peaks over Chromosomes
> files=getSampleFiles()
> peak=readPeakFile(files[[4]])
> peak
GRanges object with 1331 ranges and 2 metadata columns:
seqnames ranges strand | V4 V5
|
[1] chr1 [ 815092, 817883] * | MACS_peak_1 295.76
[2] chr1 [1243287, 1244338] * | MACS_peak_2 63.19
[3] chr1 [2979976, 2981228] * | MACS_peak_3 100.16
[4] chr1 [3566181, 3567876] * | MACS_peak_4 558.89
[5] chr1 [3816545, 3818111] * | MACS_peak_5 57.57
... ... ... ... ... ... ...
[1327] chrX [135244782, 135245821] * | MACS_peak_1327 55.54
[1328] chrX [139171963, 139173506] * | MACS_peak_1328 270.19
[1329] chrX [139583953, 139586126] * | MACS_peak_1329 918.73
[1330] chrX [139592001, 139593238] * | MACS_peak_1330 210.88
[1331] chrY [ 13845133, 13845777] * | MACS_peak_1331 58.39
---
seqlengths:
chr1 chr10 chr11 chr12 chr13 chr14 ... chr6 chr7 chr8 chr9 chrX chrY
NA NA NA NA NA NA ... NA NA NA NA NA NA
> covplot(peak, weightCol="V5")
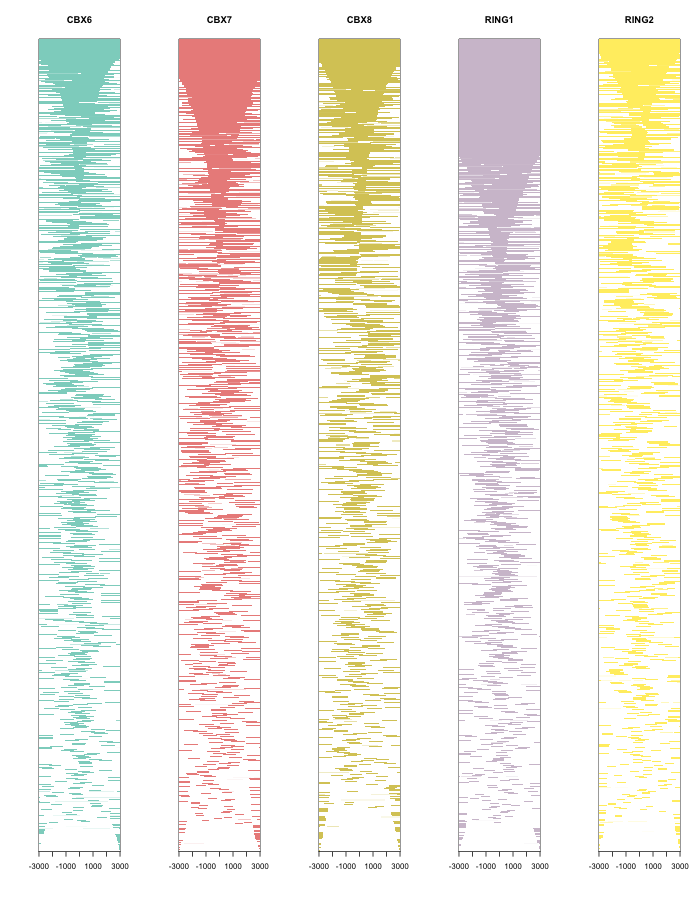
Heatmap of ChIP binding to TSS regions
require(TxDb.Hsapiens.UCSC.hg19.knownGene)
txdb <- TxDb.Hsapiens.UCSC.hg19.knownGene
peakHeatmap(files, weightCol="V5", TxDb=txdb, upstream=3000, downstream=3000, color=rainbow(length(files)))
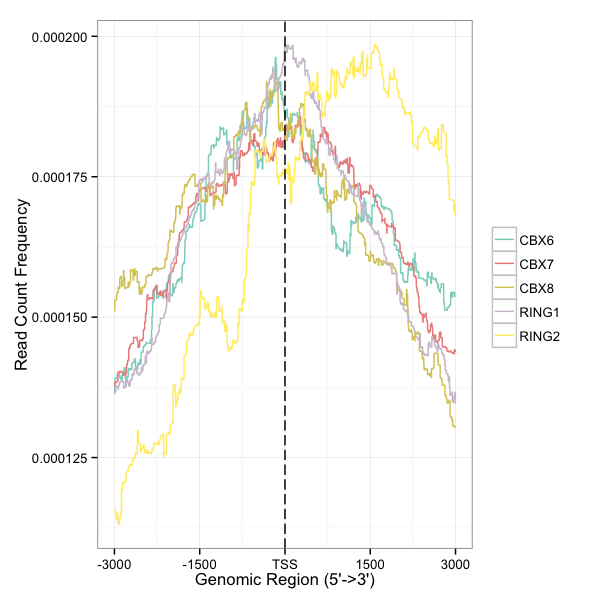
Average Profile of ChIP peaks binding to TSS region
plotAvgProf(files, TranscriptDb=txdb, weightCol="V5", upstream=3000, downstream=3000)
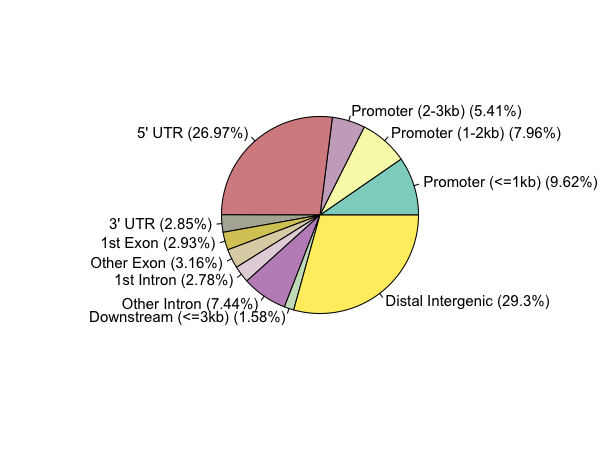
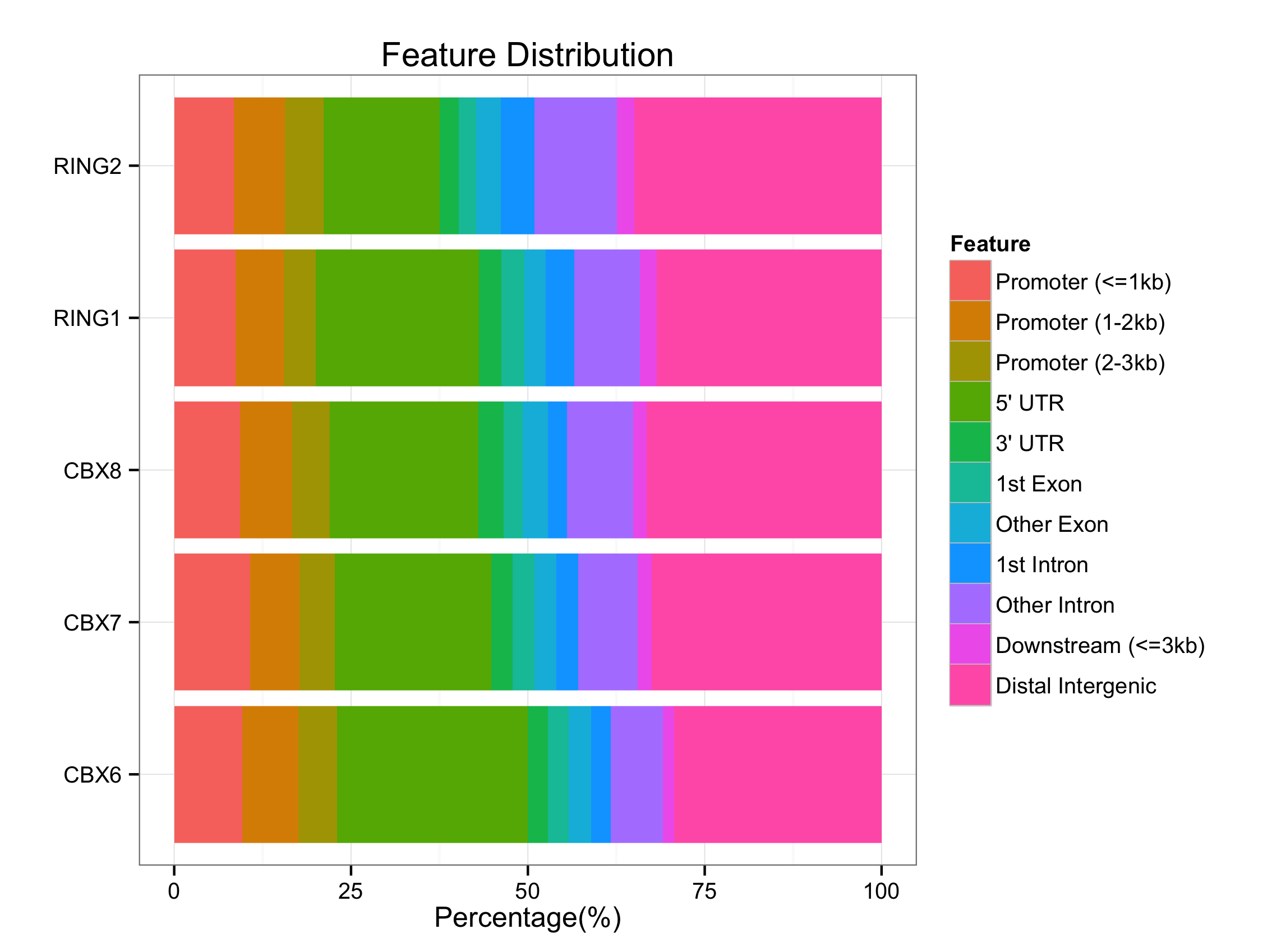
Genomic Annotation
peakAnnoList=lapply(files, annotatePeak)
plotAnnoPie(peakAnnoList[[1]])
plotAnnoBar(peakAnnoList)
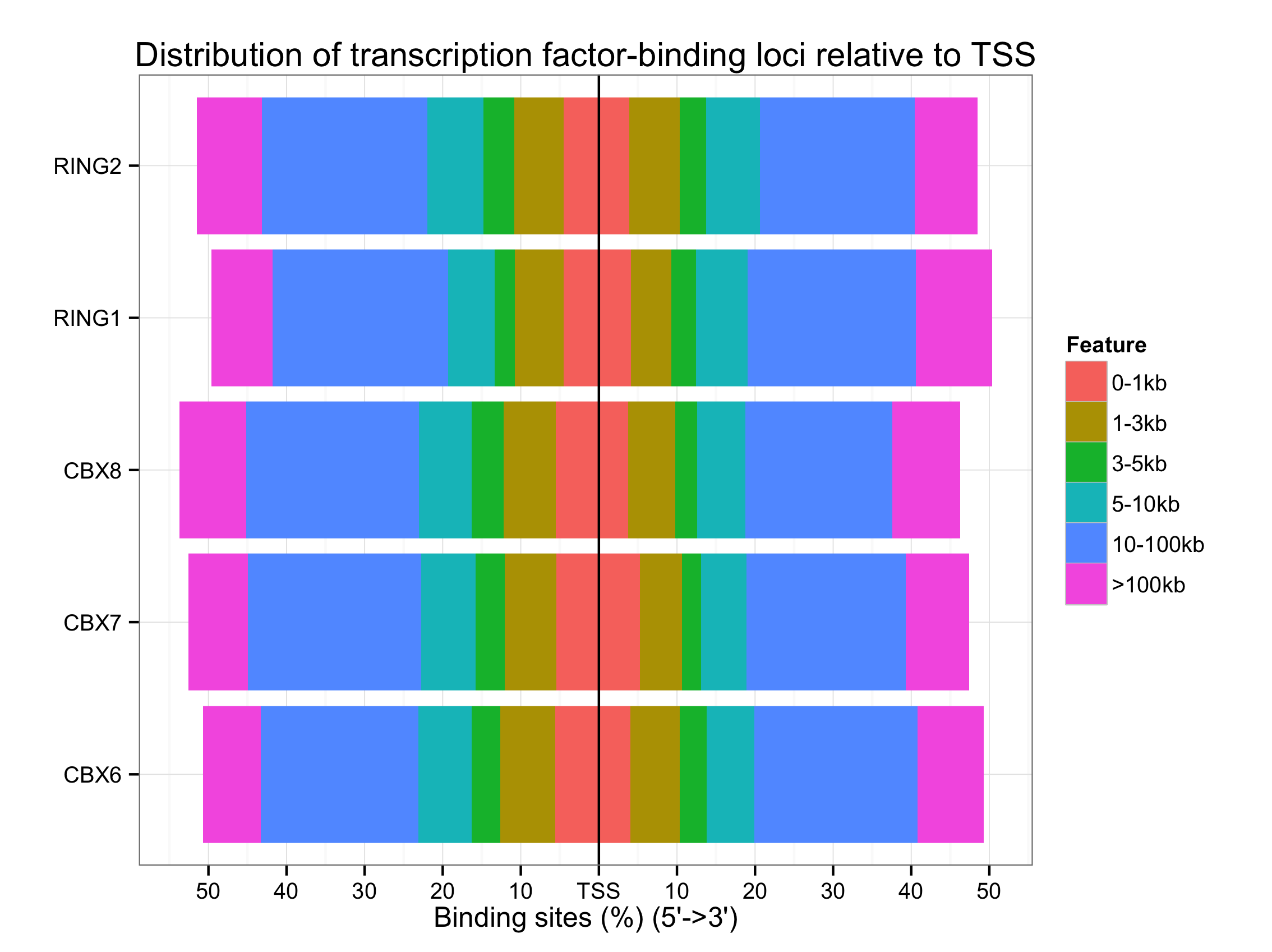
Distance to TSS
plotDistToTSS(peakAnnoList)
Overlap of peak sets
genes= lapply(peakAnnoList, function(i) as.data.frame(i)$geneId)
vennplot(genes)
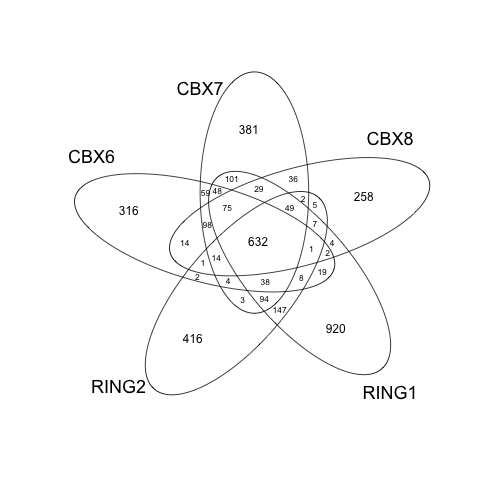
In the future version, ChIPseeker will support statistical comparison among ChIP peak sets, and incorporate open access database GEO for users to compare their own dataset to those deposited in database. Significant overlap among peak sets can be used to infer cooperative regulation. This feature will soon be available.
Citation
Yu G, Wang LG and He QY*. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015, 31(14):2382-2383.