using GOSemSim to rank proteins obtained by co-IP
Co-IP is usually used to identified interactions among specific proteins. It is widely used in detecting protein complex. Unfortunately, an identified protein may not be an interactor, and sometimes can be a background contaminant. Ranking proteins can help us to focus a study on a few high quality candidates for subsequent interaction investigation. My R package GOSemSim has been used in many studies to investigate protein protein interaction, see 1, 2, 3, 4 and more.
In the study, Proteomic investigation of the interactome of FMNL1 in hematopoietic cells unveils a role in calcium-dependent membrane plasticity, we investigate the interactors of FMNL1 by co-IP followed by LC-MS/MS analysis. In this study, we defined functional similarity as the geometric mean of semantic similarities in molecular function and cellular component. Functional similarity is designed for measuring the strength of the relationship between each protein and its partners by considering function and location of proteins. The similarities were calculated through GOSemSim package. The distributions of the functional similarities were demonstrated in the following figure.
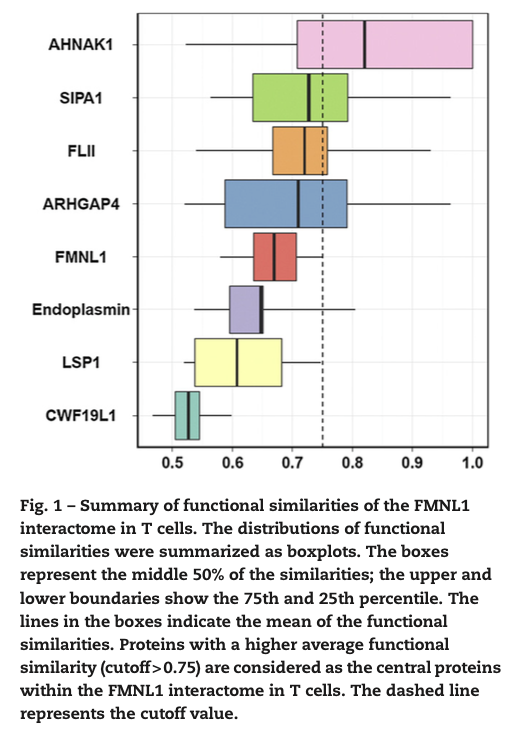
We used the average functional similarities between the protein and its interaction partners to rank the protein. Proteins, which have strong relationship in function and location among the proteins within the interactome, were essential for the interactome to exert their functions. AHNAK1 has strong spatial and temporal interaction with other proteins and was selected for functional study. AHNAK1 had not been identified as an interaction partner of FMNL1, and we confirmed that it interacts at its C-terminus with N-terminal region of FMNL1. Previous study had reported AHNAK1 is calcium-dependent, and our study found that FMNL1 enhances ionophore-mediated calcium influx by inducing translocation of AHNAK1. This point to a new function of FMNL1 as a modulator in calcium-dependent cytoskeletal membrane processes.
Citation
Yu G†, Li F†, Qin Y, Bo X*, Wu Y and Wang S*. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics. 2010, 26(7):976-978.