News of ggtree
A new version of ggtree that works with ggplot2 (version >= 2.0.0) is now availabel.
new layers
Some functions, add_legend, hilight, annotation_clade and annotation_clade2 were removed. Instead we provide layer functions, geom_treescale, geom_hilight and geom_cladelabel. You can use + operator to add layers using these layer functions.
In addtion, we provide geom_point2, geom_text2 and geom_segment2 which works exactly as geom_point, geom_text and geom_segment except they allow ggtree users to do subsetting.
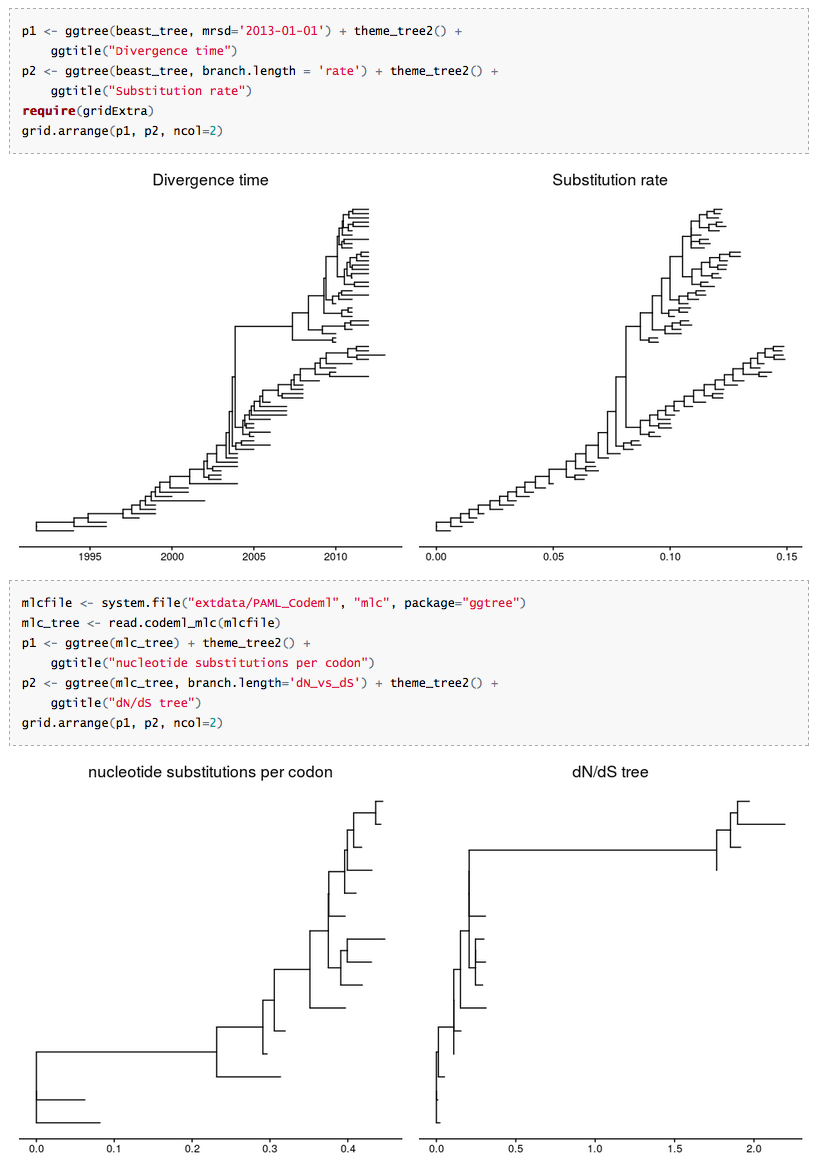
rescale tree
Most of the phylogenetic trees are scaled by evolutionary distance (substitution/site), in ggtree we can re-scale a phylogenetic tree by any numerical variable inferred by evolutionary analysis. For example using substitution rate to scale a time-scaled tree inferred by BEAST, or using dN/dS to re-scale a tree analyzed by CodeML.
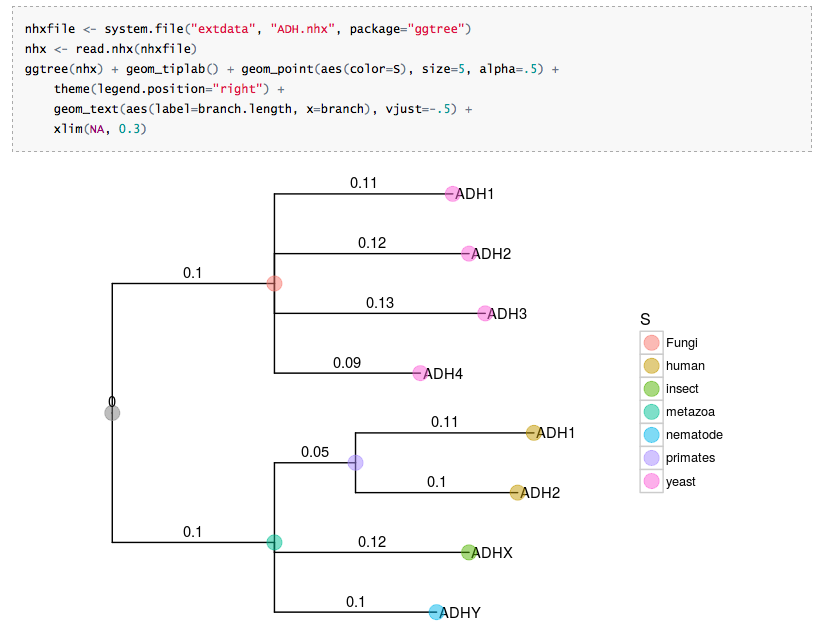
New Hampshire eXtended format (NHX) supported
This is a feature request from ggtree user. The NHX format is also commonly used in phylogenetic software (e.g. PHYLODOG, RevBayes, etc).
The original vignette was too long and I split them into several short ones. These vignettes should be more easy to follow. You can access the vignette via http://guangchuangyu.github.io/ggtree/. More examples and detail explanation were provided. If there are something that I didn’t explain well or if you have suggestions for changes or improvements, submit an issue, or (even better) fork ggtree, make modifications and submit a pull request.
I appreciate your help in improving ggtree.
- Repo: https://github.com/GuangchuangYu/ggtree
- vignettes
Citation
G Yu, DK Smith, H Zhu, Y Guan, TTY Lam*. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution. doi:10.1111/2041-210X.12628.