ggtree supports phylip tree format
Phylip is also a widely used tree format, which contains taxa sequences with Newick tree text.
In ggtree, we can use read.phylip() function to parse the file and use ggtree() to visualize the tree.
library(ggtree) phyfile <- system.file(“extdata”, “sample.phy”, package=“ggtree”) phylip <- read.phylip(phyfile) phylip
## 'phylip' S4 object that stored information of
## '/Users/guangchuangyu/Library/R/3.2/library/ggtree/extdata/sample.phy'.
##
## ...@ tree:
## Phylogenetic tree with 15 tips and 13 internal nodes.
##
## Tip labels:
## K, N, D, L, J, G, ...
##
## Unrooted; no branch lengths.
##
## with sequence alignment available (15 sequences of length 2148)
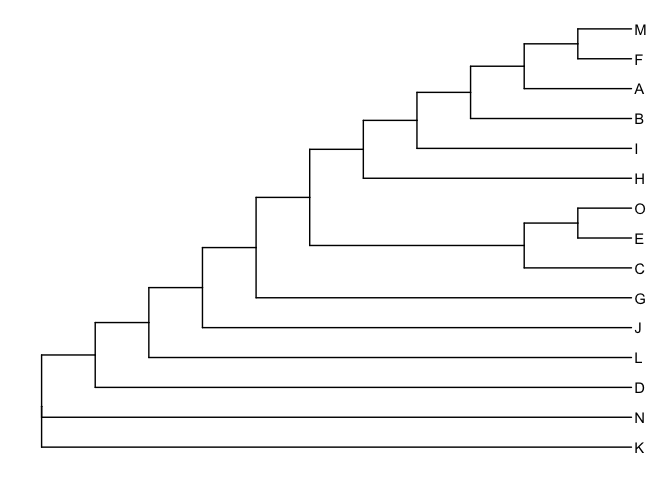
ggtree(phylip) + geom_tiplab()
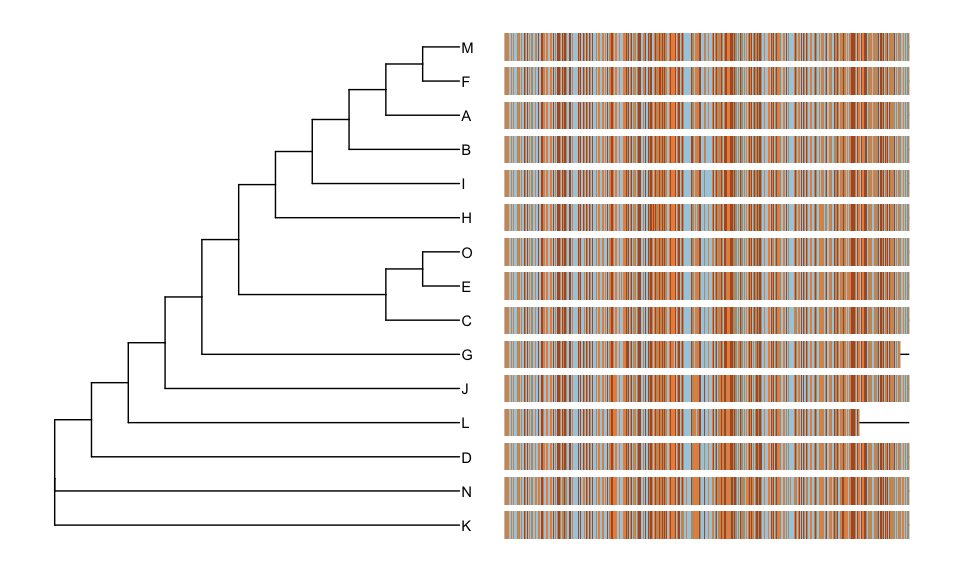
User can view the sequence alignment with the tree via msaplot()
function.
msaplot(phylip, offset=1)
Citation
G Yu, DK Smith, H Zhu, Y Guan, TTY Lam*. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution. doi:10.1111/2041-210X.12628.