parsing BED coordinates
In supplemental file of ChIPseeker paper, I compare distances to TSS reported by several ChIP annotation software, including ChIPseeker, ChIPpeakAnno, HOMER and PeakAnalyzer.
Although I found that the chromStart positions in HOMER output have a +1 shift compare to other software, I did not realize this issue since all other software are consistent.
Until recently, I found BAM, BCF, BED and PSL formats are using the 0-based coordinate system, while SAM, VCF, GFF and Wiggle formats are using the 1-based coordinate system.
For BED file format, we can refer to http://asia.ensembl.org/info/website/upload/bed.html.
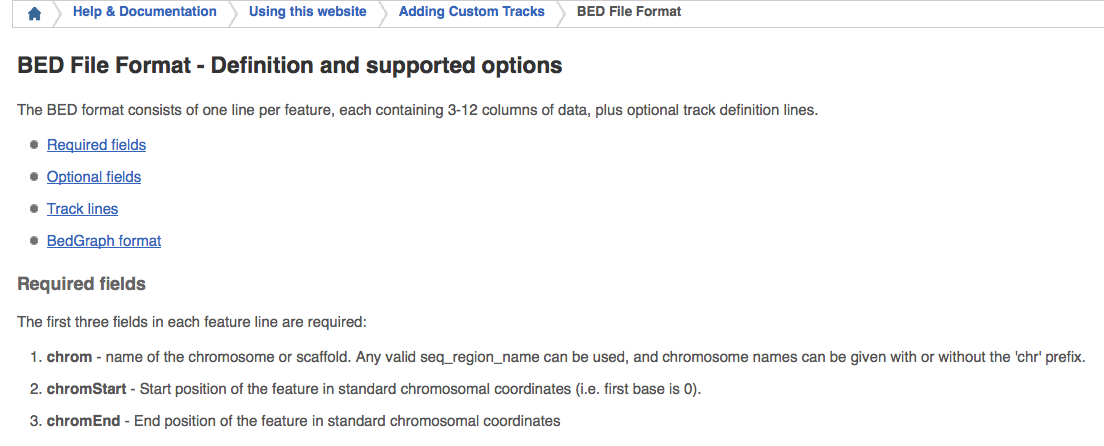
In addition, the 0-based coordinate system is specified by a
half-closed-half-open interval. For example, the first 100 bases of a
chromosome are defined as [0, 100), that span the bases numbered 0-99.
While the 1-based coordinate system is specified by a closed interval,
for example the above region is [1, 100].
Most of the software for ChIP annotation doesn’t considered this issue
when annotating peak (0-based) to transcript (1-based). To my knowledge,
only HOMER consider this issue. After I figure this out, I have updated
ChIPseeker (version
>= 1.4.3) to fix the issue.
Please bare in mind that chromStart in ChIPseeker has +1bp shift compare to the number recorded in BED file.
Citation
Yu G, Wang LG and He QY*. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015, 31(14):2382-2383.