Annotate a phylogenetic tree with insets
ggtree implemented a function, subview, that can add subplots on a
ggplot2 object. It had successful applied to plot pie graphs on map.
set.seed(2016-01-04)
tr <- rtree(30)
tr <- groupClade(tr, node=45)
p <- ggtree(tr, aes(color=group)) + geom_tippoint()
cpos <- get_clade_position(p, node=45)
p1 <- p + geom_hilight(node=45)
p2 <- with(cpos, p+xlim(xmin, xmax*1.01)+ylim(ymin, ymax))
with(cpos, subview(p2+geom_tiplab(), p1+theme_transparent(), x=xmin+(xmax-xmin)*.15, y=ymin+(ymax-ymin)*.85))
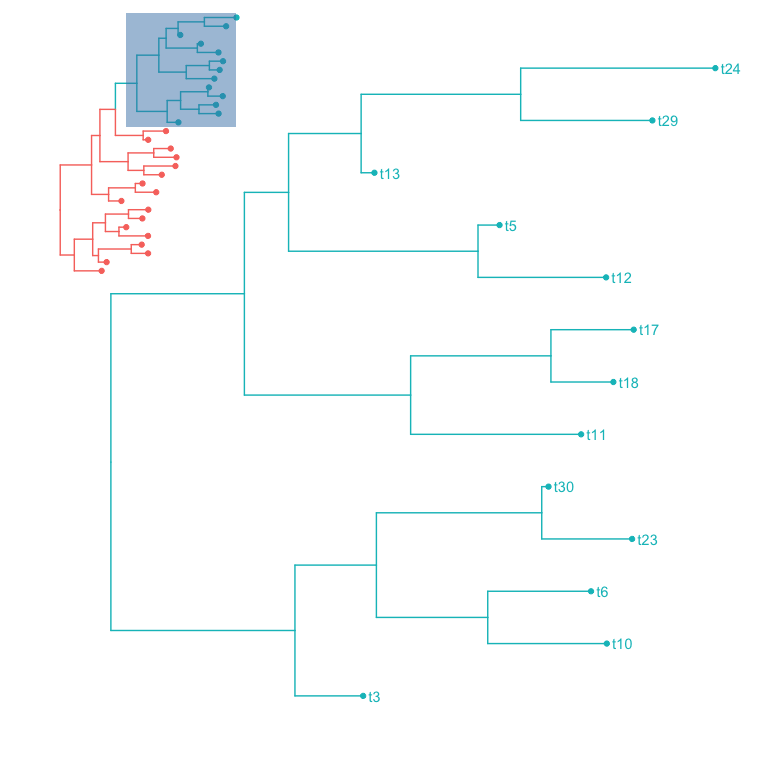
To make it more easy to use subview function for annotating taxa with
subplots, ggtree provides a function, inset, for adding subplots to
a phylogenetic tree. The input is a tree graphic object and a named list
of ggplot graphic objects (can be any kind of charts), these objects
should named by node numbers. To facilitate adding bar and pie charts
(e.g. summarized stats of results from ancestral reconstruction) to
phylogenetic tree, ggtree provides nodepie and nodebar functions
to create a list of pie or bar charts.
Annotate with bar charts
set.seed(2015-12-31)
tr <- rtree(15)
p <- ggtree(tr)
a <- runif(14, 0, 0.33)
b <- runif(14, 0, 0.33)
c <- runif(14, 0, 0.33)
d <- 1 - a - b - c
dat <- data.frame(a=a, b=b, c=c, d=d)
## input data should have a column of `node` that store the node number
dat$node <- 15+1:14
## cols parameter indicate which columns store stats (a, b, c and d in this example)
bars <- nodebar(dat, cols=1:4)
inset(p, bars)
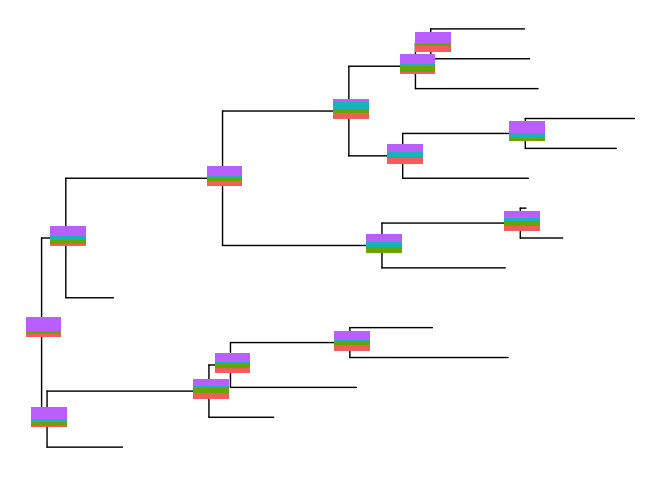
The sizes of the insets can be ajusted by the paramter width and
height.
inset(p, bars, width=.03, height=.06)
Users can set the color via the parameter color. The x position can
be one of ‘node’ or ‘branch’ and can be adjusted by the parameter
hjust and vjust for horizontal and vertical adjustment respecitvely.
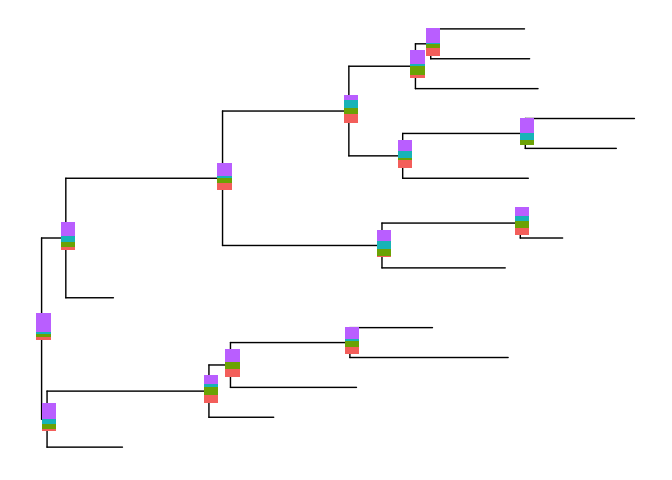
bars2 <- nodebar(dat, cols=1:4, position='dodge',
color=c(a='blue', b='red', c='green', d='cyan'))
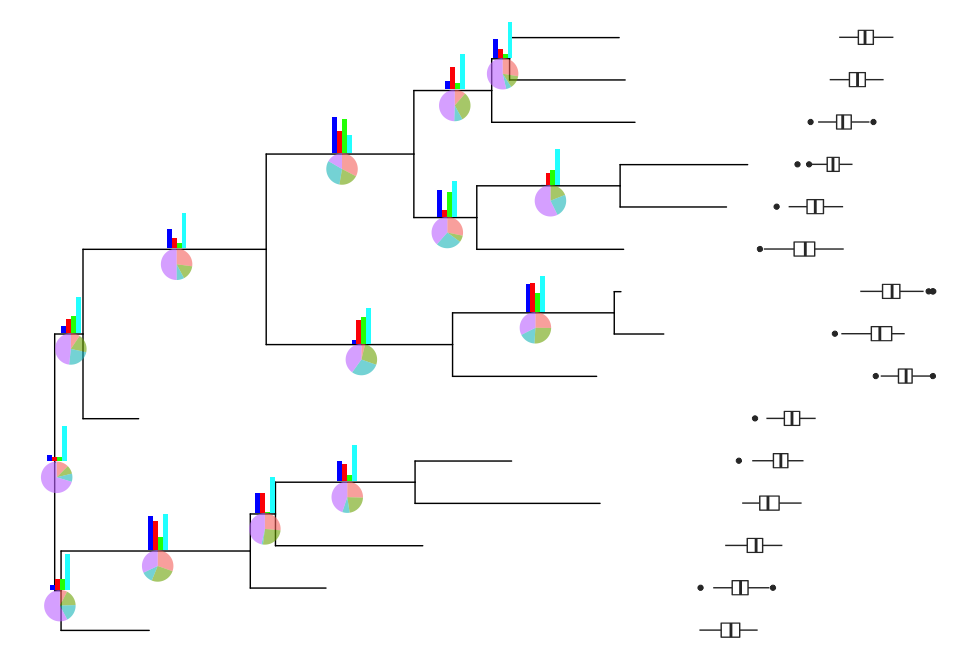
p2 <- inset(p, bars2, x='branch', width=.03, vjust=-.3)
print(p2)
Annotate with pie charts
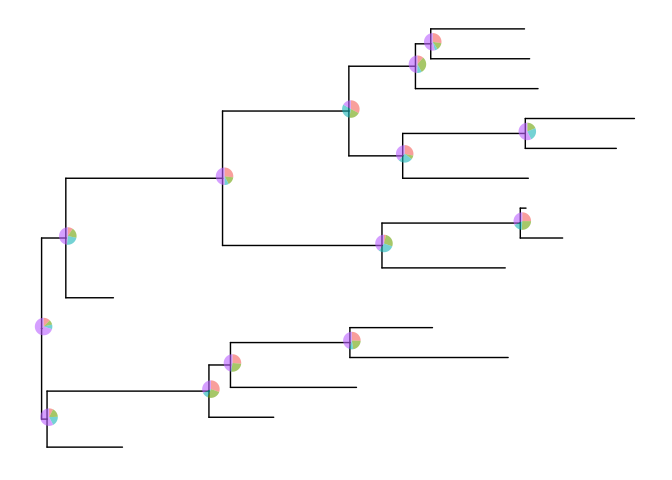
Similarly, users can use nodepie function to generate a list of pie
charts and place these charts to annotate corresponding nodes. Both
nodebar and nodepie accepts parameter alpha to allow transparency.
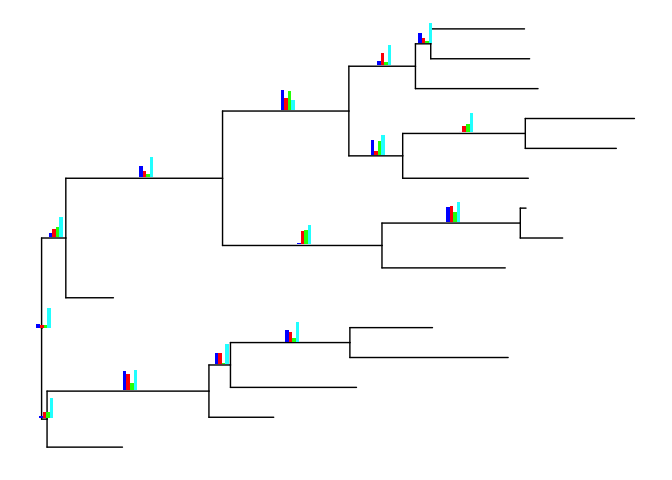
pies <- nodepie(dat, cols=1:4, alpha=.6)
inset(p, pies)
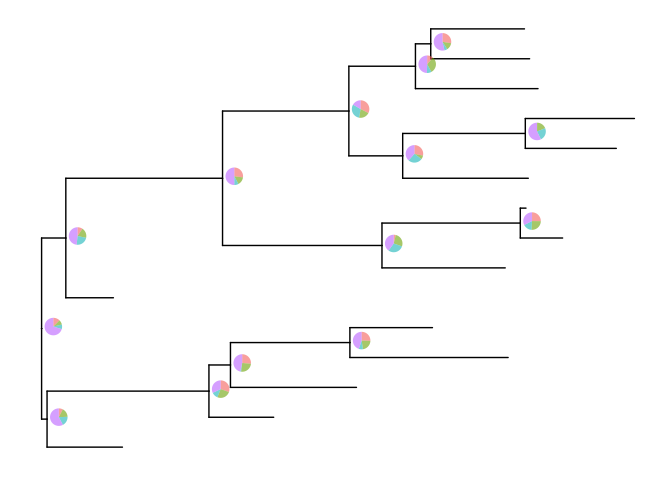
inset(p, pies, hjust=-.06)
Annotate with other types of charts
The inset function accepts a list of ggplot graphic objects and these
input objects are not restricted to pie or bar charts. They can be any
kinds of charts and hybrid of these charts.
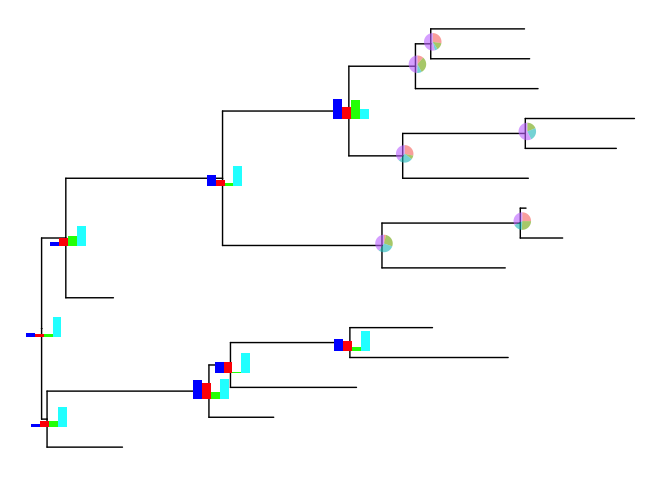
pies_and_bars <- bars2
pies_and_bars[9:14] <- pies[9:14]
inset(p, pies_and_bars)
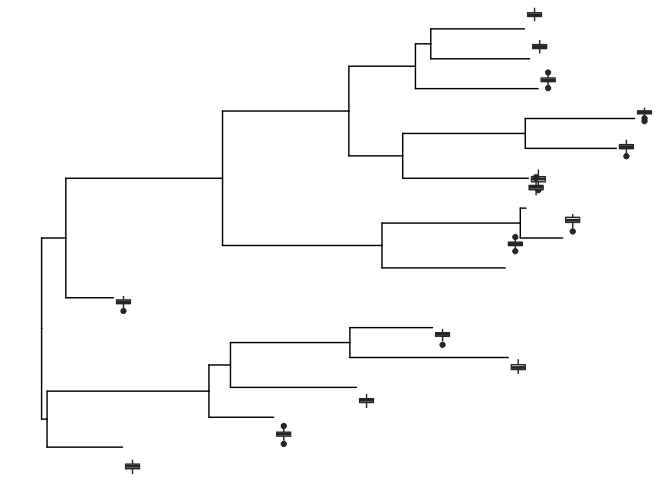
d <- lapply(1:15, rnorm, n=100)
ylim <- range(unlist(d))
bx <- lapply(d, function(y) {
dd <- data.frame(y=y)
ggplot(dd, aes(x=1, y=y))+geom_boxplot() + ylim(ylim) + theme_inset()
})
names(bx) <- 1:15
inset(p, bx, width=.03, height=.1, hjust=-.05)
After annotating with insets, users can further annotate the tree with another layer of insets.
p2 <- inset(p, bars2, x='branch', width=.03, vjust=-.4)
p2 <- inset(p2, pies, x='branch', vjust=.4)
bx2 <- lapply(bx, function(g) g+coord_flip())
inset(p2, bx2, width=.2, height=.03, vjust=.04, hjust=p2$data$x[1:15]-4) + xlim(NA, 4.5)
Citation
G Yu, DK Smith, H Zhu, Y Guan, TTY Lam*. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution. doi:10.1111/2041-210X.12628.