convert graphic object to tree object using treeio
I have splitted ggtree to 2 packages, treeio and ggtree. Now ggtree is mainly focus on visualization and annotation, while treeio focus on parsing and exporting tree files. Here is a welcome message from treeio that you can convert ggtree output to tree object which can be exported as newick or nexus file if you want.
Thanks to ggplot2, output of ggtree is actually a ggplot object. The ggtree object can be rendered as graph by print method, but internally it is an object that contains data. treeio defines as.phylo and as.treedata to convert ggtree object to phylo or treedata object.
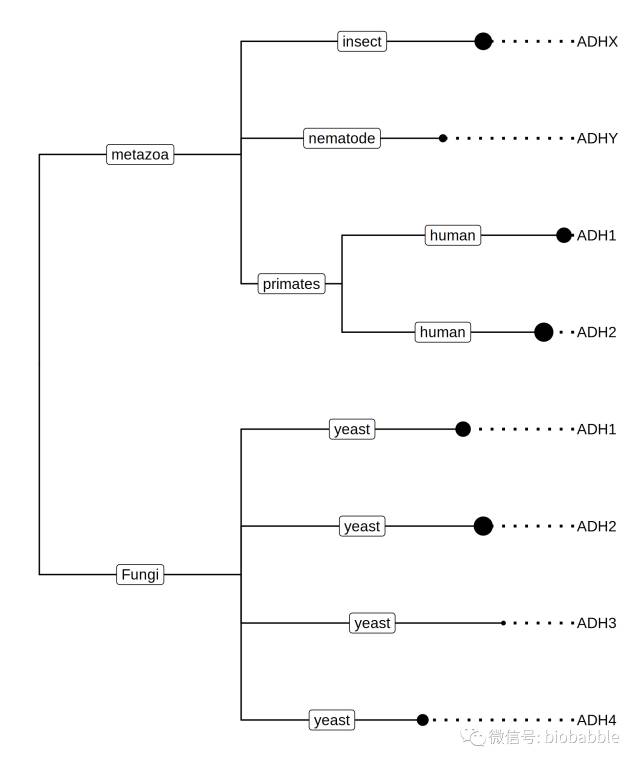
require(ggtree) nhxfile <- system.file(“extdata/NHX”, “ADH.nhx”, package=“treeio”) nhx <- read.nhx(nhxfile) p <- ggtree(nhx)
After parsing the NHX file via read.nhx function, we can visualize it using ggtree, and the output ggtree object can be converted back as a phylo object using as.phylo method:
as.phylo(p)
Phylogenetic tree with 8 tips and 4 internal nodes.
Tip labels: ADH2, ADH1, ADHY, ADHX, ADH4, ADH3, …
Rooted; includes branch lengths.
The output phylo object contains the tree structure.
If we want to also save associated annotation data, we can use as.treedata method:
as.treedata(p)
‘treedata’ S4 object that stored information of ‘’.
…@ tree: Phylogenetic tree with 8 tips and 4 internal nodes.
Tip labels:
ADH2, ADH1, ADHY, ADHX, ADH4, ADH3, …
Rooted; includes branch lengths.
with the following features available: ‘S’, ‘D’, ‘B’.
In ggtree, we can use %<+% operator to attach user’s own data to graphic object and then use it to annotate the tree. as.treedata can also export these attached data to the tree object.
d = data.frame(label=as.phylo(nhx)$tip.label, trait=abs(rnorm(Ntip(nhx))))
p = p %<+% d
as.treedata(p)
‘treedata’ S4 object that stored information of ‘’.
…@ tree:
Phylogenetic tree with 8 tips and 4 internal nodes.
Tip labels:
ADH2, ADH1, ADHY, ADHX, ADH4, ADH3, …
Rooted; includes branch lengths.
with the following features available: ‘S’, ‘D’, ‘B’, ‘trait’.
Here as an example, the trait variable was attached and exported to the tree object.
This trait variable can also be used in tree annotation.
x <- as.treedata(p)
ggtree(x) + geom_tiplab(align=T, offset=.005) +
geom_tippoint(aes(size=trait)) + xlim(NA, 0.28) +
geom_label(aes(x=branch, label=S))
We can visualize the tree by ggtree(tree_object) and we can convert the ggtree object back to tree object via as.treedata(ggtree_object).
If you lost your tree file but have the ggtree object saved, you can convert it back to tree object and export the tree object to newick or nexus file.