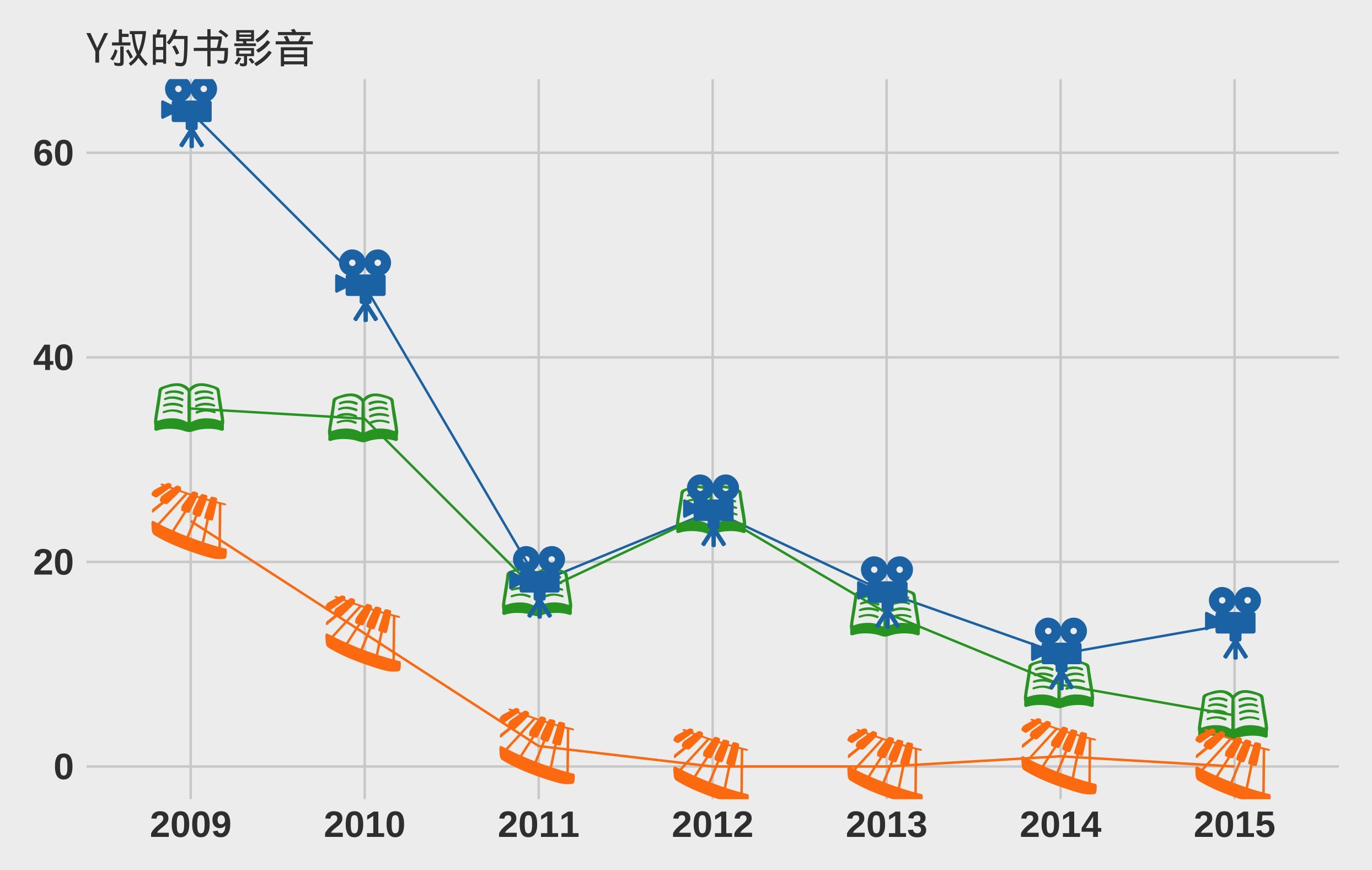
I extended the subview function to support embed image file in a ggplot object.
set.seed(123)
d = data.frame(x=rnorm(10), y=rnorm(10))
imgfile <- tempfile(, fileext=".png")
download.file("https://avatars1.githubusercontent.com/u/626539?v=3&u=e731426406dd3f45a73d96dd604bc45ae2e7c36f&s=140",
destfile=imgfile, mode='wb')
p = ggplot(d, aes(x, y))
subview(p, imgfile, x=d$x[1], y=d$y[1]) + geom_point(size=5)