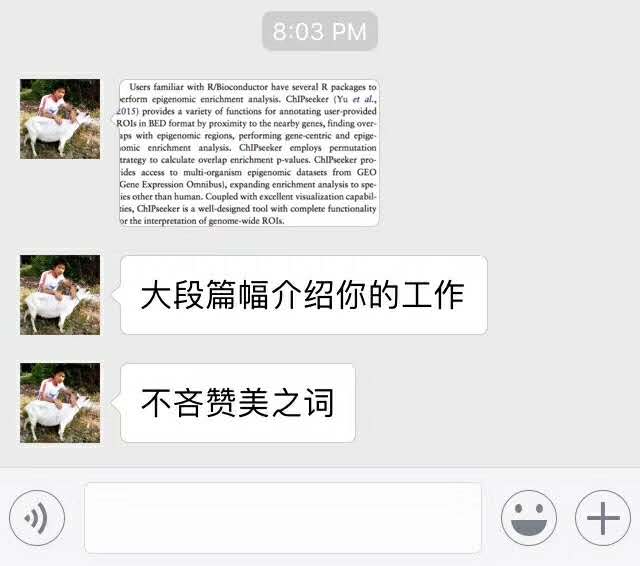
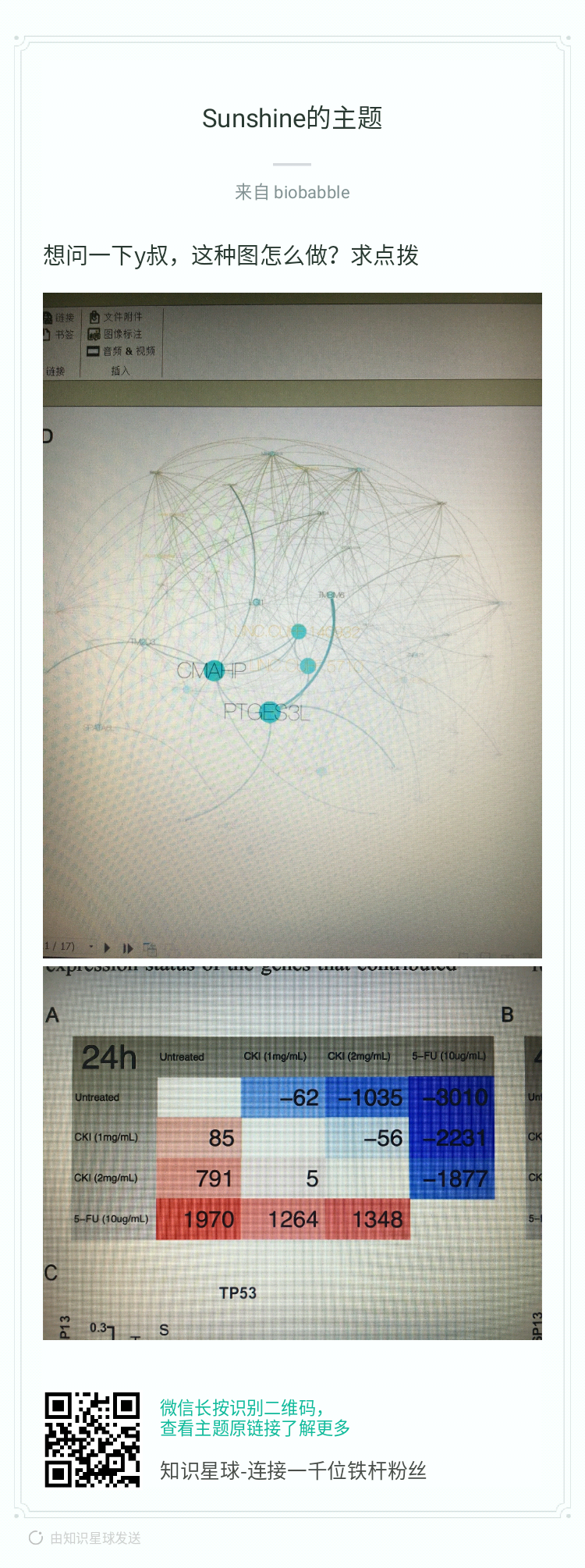
小伙伴发来这个文章中的图,想要实现类似的图,用变量给axis text上色,并生成legend:
这个可以说ggplot2是不支持的,aes映射不会被应用于axis上,而theme也不支持aes映射,你只能自己手动搞个color vector传给theme来上色,但这无法生成legend。
我只能打开脑洞,legend借助于额外的图层来生成,但这个额外的图层又不是我们想要在图形上展示的,这又是个无米之炊,我能想到的就是让图层透明,实际有,但你看不到,当然这样自动生成的legend也会看不到,但legend可以后面再修改,于是这不支持的事情,就通过变通变得可能:
require(ggplot2)
set.seed(2017-07-04)
d <- data.frame(x = letters[1:5], y = rnorm(5),
group = sample(c("Control", "Treatment"), 5, replace=TRUE),
type = sample(LETTERS[1:2], 5, replace=TRUE)
)
p <- ggplot(d, aes(x,y)) +
geom_col(aes(fill=group))
这个代码很好懂,画一个柱状图,按group上色。