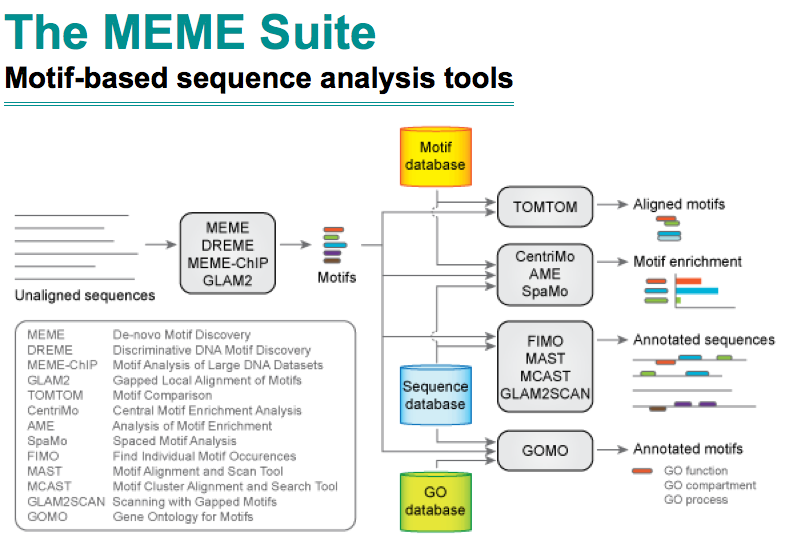
showtext is a neat solution to use various types of fonts in R graphs and make it easy to use funny fonts. With showtext, we can draw phylogenetic tree with different types of fonts even with symbolic/icon fonts.
require(showtext)
font.add.google("Gochi Hand", "gochi")
font.add.google("Rock Salt", "rock")
link = "http://img.dafont.com/dl/?f=wm_people_1";
download.file(link, "wmpeople1.zip", mode = "wb");
unzip("wmpeople1.zip");
font.add("wmpeople1", "wmpeople1.TTF");
link = "http://img.dafont.com/dl/?f=emoticons";
download.file(link, "emoticons.zip", mode = "wb");
unzip("emoticons.zip");
font.add("emoticons", "emoticons.ttf");
showtext.auto()
set.seed(2015-05-14)
library(ggplot2)
library(ggtree)
tree <- rtree(30)
ggtree(tree, color="darkgreen") + geom_tiplab(family="gochi") + theme_classic() +
theme(axis.text.x=element_text(size=rel(4), family="emoticons", color="firebrick")) +
scale_y_continuous(breaks=seq(0, 30, 4), labels=letters[c(12:17, 20, 21)]) +
theme(axis.text.y=element_text(size=rel(4), family="wmpeople1", color="#FDAC4F")) +
annotate("text", x=2, y=18, label="Have fun with ggtree!",
family="rock", angle=30, size=12, color="steelblue")