ChIPseeker: ChIP peak Annotation, Comparison, and Visualization
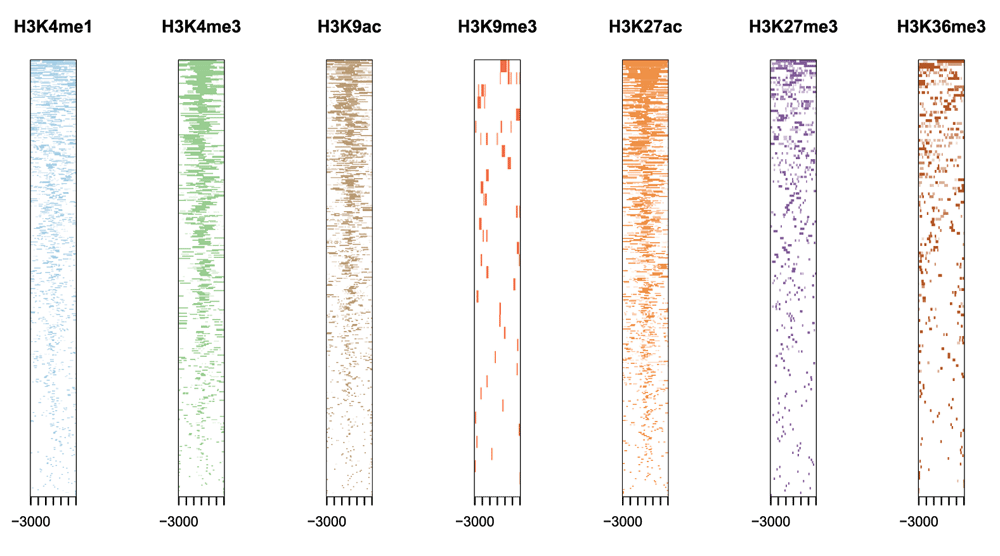
This package implements functions to retrieve the nearest genes around the peak, annotate genomic region of the peak, statstical methods for estimate the significance of overlap among ChIP peak data sets, and incorporate GEO database for user to compare their own dataset with those deposited in database. The comparison can be used to infer cooperative regulation and thus can be used to generate hypotheses. Several visualization functions are implemented to summarize the coverage of the peak experiment, average profile and heatmap of peaks binding to TSS regions, genomic annotation, distance to TSS, and overlap of peaks or genes.
ChIPseeker is released within the Bioconductor project and the source code is hosted on GitHub.
Citation
Please cite the following article when using ChIPseeker:
Yu G, Wang LG and He QY*. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics, 2015, 31(14):2382-2383.
Installation
Install ChIPseeker is easy, follow the guide on the Bioconductor page:
## try http:// if https:// URLs are not supported
source("https://bioconductor.org/biocLite.R")
## biocLite("BiocUpgrade") ## you may need this
biocLite("ChIPseeker")
Overview
Annotation
- retrieve the nearest genes around the peak
- annotate genomic region of the peak
Comparison
- estimate the significance of overlap among ChIP peak data sets
- incorporate GEO database for users to compare their own dataset with those deposited in the database
Visualization
- summarize the coverage of the peak experiment
- average profile and heatmap of peaks binding to TSS regions
- genomic annotation
- distance to TSS
- overlap of peaks or genes
Feedback
- Please make sure you have followed the important guide before posting any issue/question
- For bugs or feature requests, please post to github issue
- For user questions, please post to Bioconductor support site or Biostars
- Join the group chat in and