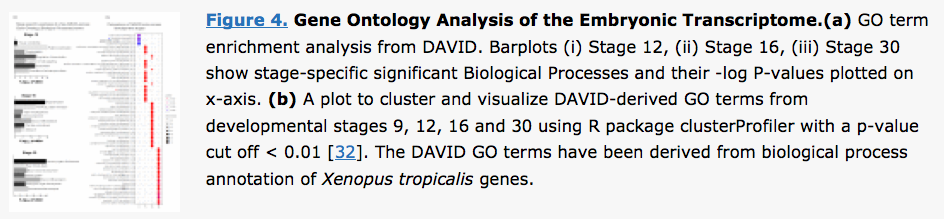
clusterProfiler was
used to visualize DAVID
results in a paper published in BMC
Genomics.
Some users told me that they may want to use DAVID at some
circumstances. I think it maybe a good idea to make clusterProfiler
supports DAVID, so that
DAVID users can use
visualization functions provided by
clusterProfiler.
require(DOSE)
require(clusterProfiler)
data(geneList)
gene = names(geneList)[abs(geneList) > 2]
david = enrichDAVID(gene = gene, idType="ENTREZ_GENE_ID",
listType="Gene", annotation="KEGG_PATHWAY")
> summary(david)
ID Description GeneRatio BgRatio pvalue
hsa04110 hsa04110 Cell cycle 11/68 125/5085 4.254437e-06
hsa04114 hsa04114 Oocyte meiosis 10/68 110/5085 1.119764e-05
hsa03320 hsa03320 PPAR signaling pathway 7/68 69/5085 2.606715e-04
p.adjust qvalue geneID
hsa04110 0.0003998379 NA 9133/4174/890/991/1111/891/7272/8318/4085/983/9232
hsa04114 0.0005261534 NA 9133/5241/51806/3708/991/891/4085/983/9232/6790
hsa03320 0.0081354974 NA 4312/2167/5346/5105/3158/9370/9415
Count
hsa04110 11
hsa04114 10
hsa03320 7
There are only 5085 human genes annotated by KEGG, this is due to
out-of-date DAVID data.
Continue reading