leading edge analysis
leading edge and core enrichment
Leading edge analysis reports Tags to indicate the percentage of genes contributing to the enrichment score, List to indicate where in the list the enrichment score is attained and Signal for enrichment signal strength.
It would also be very interesting to get the core enriched genes that contribute to the enrichment.
Now DOSE, clusterProfiler and ReactomePA all support leading edge analysis and report core enriched genes.
library(DOSE) data(geneList) x <- gseDO(geneList)
## [1] "preparing geneSet collections..."
## [1] "GSEA analysis..."
## [1] "leading edge analysis..."
## [1] "done..."
head(x)
## ID Description setSize
## DOID:1492 DOID:1492 eye and adnexa disease 459
## DOID:5614 DOID:5614 eye disease 450
## DOID:0060037 DOID:0060037 developmental disorder of mental health 315
## DOID:374 DOID:374 nutrition disease 313
## DOID:5679 DOID:5679 retinal disease 299
## DOID:654 DOID:654 overnutrition 298
## enrichmentScore NES pvalue p.adjust qvalues
## DOID:1492 -0.3105160 -1.379586 0.001231527 0.03329506 0.02257366
## DOID:5614 -0.3125247 -1.386602 0.001243781 0.03329506 0.02257366
## DOID:0060037 -0.3407349 -1.473028 0.001307190 0.03329506 0.02257366
## DOID:374 -0.3421127 -1.477222 0.001310616 0.03329506 0.02257366
## DOID:5679 -0.3676313 -1.582707 0.001333333 0.03329506 0.02257366
## DOID:654 -0.3490198 -1.500301 0.001340483 0.03329506 0.02257366
## rank leading_edge
## DOID:1492 1793 tags=22%, list=14%, signal=19%
## DOID:5614 1768 tags=22%, list=14%, signal=19%
## DOID:0060037 2313 tags=24%, list=19%, signal=20%
## DOID:374 1464 tags=22%, list=12%, signal=20%
## DOID:5679 1768 tags=24%, list=14%, signal=21%
## DOID:654 1464 tags=21%, list=12%, signal=19%
## core_enrichment
## DOID:1492 3371/3082/5914/2878/4153/3791/23247/1543/80184/6750/1958/2098/7450/596/9187/2034/482/948/1490/1280/3931/5737/4314/4881/2261/3426/187/629/6403/7042/6785/7507/2934/5176/4060/1277/7078/5950/2057/727/10516/4311/2247/1295/358/10203/2192/582/10218/57125/3485/585/1675/6310/2202/4313/2944/4254/3075/1501/2099/3480/4653/1195/6387/3305/1471/857/4016/1909/4053/6678/1296/7033/4915/55812/1191/5654/10631/2152/2697/7043/2952/6935/2200/3572/7177/7031/3479/2006/10451/9370/771/3117/125/652/4693/5346/1524
## DOID:5614 3082/5914/2878/4153/3791/23247/1543/80184/6750/1958/2098/7450/596/9187/2034/482/948/1490/1280/3931/5737/4314/4881/2261/3426/187/629/6403/7042/6785/7507/2934/5176/4060/1277/7078/5950/2057/727/10516/4311/2247/1295/358/10203/2192/582/10218/57125/3485/585/1675/6310/2202/4313/2944/4254/3075/1501/2099/3480/4653/6387/3305/1471/857/4016/1909/4053/6678/1296/7033/4915/55812/1191/5654/10631/2152/2697/7043/2952/6935/2200/3572/7177/7031/3479/2006/10451/9370/771/3117/125/652/4693/5346/1524
## DOID:0060037 154/1760/9732/7337/5175/6532/4763/54806/9759/6326/1499/7157/221037/627/2908/3399/2571/3082/23503/3791/51265/27347/55650/596/3067/51185/7552/22829/23426/324/5021/4885/7248/8910/8604/3397/4208/3400/26470/553/3953/6812/64221/80208/5172/9037/3952/477/93664/3625/2944/6925/6594/7102/3908/2550/4915/4922/26960/1746/2697/6863/3913/2891/367/4128/150/7166/6505/5348/18/4129/9370/57502/4137/79083
## DOID:374 2169/1490/7840/4887/4314/595/4018/6403/590/3087/866/66036/5919/5176/3953/164656/5950/2638/2166/5243/5468/5108/10560/4023/3485/7350/3952/1149/585/1513/3489/79068/4671/477/4313/3625/9369/6720/7494/2099/3480/3991/23446/6678/4915/5167/8228/165/2152/185/367/4982/3667/4128/9607/3572/150/563/1489/3479/9370/9122/5105/2167/5346/79689/5241
## DOID:5679 2878/3791/23247/80184/6750/7450/596/9187/2034/482/948/1490/1280/5737/4314/4881/3426/187/629/6403/6785/2934/5176/7078/5950/727/10516/4311/2247/1295/358/10203/582/10218/57125/585/1675/6310/2202/4313/2944/4254/3075/2099/3480/4653/6387/1471/857/4016/1909/4053/6678/1296/4915/55812/1191/5654/10631/2697/2952/6935/2200/3479/2006/10451/9370/771/652/4693/5346/1524
## DOID:654 2169/1490/7840/4887/4314/595/4018/6403/590/3087/866/66036/5919/5176/3953/5950/2638/2166/5243/5468/4023/3485/7350/3952/1149/585/1513/3489/79068/4671/477/4313/3625/9369/6720/7494/2099/3480/3991/6678/4915/5167/8228/165/2152/185/367/4982/3667/4128/9607/3572/150/563/1489/3479/9370/9122/5105/2167/5346/79689/5241
cnetplot for core enriched genes
With core enriched genes reported, cnetplot function for GSEA result is also implemented.
setReadable
To make the plot more readable, we can use setReadable to convert Entrez ID to Gene Symbol.
y <- setReadable(x, 'org.Hs.eg.db')
head(y, 2)
## ID Description setSize
## DOID:1492 DOID:1492 eye and adnexa disease 459
## DOID:5614 DOID:5614 eye disease 450
## enrichmentScore NES pvalue p.adjust qvalues
## DOID:1492 -0.3105160 -1.379586 0.001231527 0.03329506 0.02257366
## DOID:5614 -0.3125247 -1.386602 0.001243781 0.03329506 0.02257366
## rank leading_edge
## DOID:1492 1793 tags=22%, list=14%, signal=19%
## DOID:5614 1768 tags=22%, list=14%, signal=19%
## core_enrichment
## DOID:1492 TNC/HGF/RARA/GPX3/MBL2/KDR/KIAA0556/CYP1A1/CEP290/SST/EGR1/ESD/VWF/BCL2/SLC24A1/EPAS1/ATP1B2/CD36/CTGF/COL2A1/LCAT/PTGFR/MMP3/NPR1/FGFR3/CFI/APLNR/CFB/SELP/TGFB2/ELOVL4/XPA/GSN/SERPINF1/LUM/COL1A1/TIMP3/RBP4/EPOR/C5/FBLN5/MME/FGF2/COL8A1/AQP1/CALCRL/FBLN1/BBS1/ANGPTL7/PLXDC1/IGFBP2/BBS4/CFD/ATXN1/EFEMP1/MMP2/GSTM1/KITLG/CFH/CTNND2/ESR1/IGF1R/MYOC/CLK1/CXCL12/HSPA1L/CST3/CAV1/LOXL1/EDNRA/LTBP2/SPARC/COL8A2/TFF3/NTRK2/SPATA7/CLU/HTRA1/POSTN/F3/GJA1/TGFB3/GSTT1/ZEB1/FBN1/IL6ST/TPSAB1/TFF1/IGF1/ELN/VAV3/ADIPOQ/CA12/HLA-DQA1/ADH1B/BMP4/NDP/PLIN1/CX3CR1
## DOID:5614 HGF/RARA/GPX3/MBL2/KDR/KIAA0556/CYP1A1/CEP290/SST/EGR1/ESD/VWF/BCL2/SLC24A1/EPAS1/ATP1B2/CD36/CTGF/COL2A1/LCAT/PTGFR/MMP3/NPR1/FGFR3/CFI/APLNR/CFB/SELP/TGFB2/ELOVL4/XPA/GSN/SERPINF1/LUM/COL1A1/TIMP3/RBP4/EPOR/C5/FBLN5/MME/FGF2/COL8A1/AQP1/CALCRL/FBLN1/BBS1/ANGPTL7/PLXDC1/IGFBP2/BBS4/CFD/ATXN1/EFEMP1/MMP2/GSTM1/KITLG/CFH/CTNND2/ESR1/IGF1R/MYOC/CXCL12/HSPA1L/CST3/CAV1/LOXL1/EDNRA/LTBP2/SPARC/COL8A2/TFF3/NTRK2/SPATA7/CLU/HTRA1/POSTN/F3/GJA1/TGFB3/GSTT1/ZEB1/FBN1/IL6ST/TPSAB1/TFF1/IGF1/ELN/VAV3/ADIPOQ/CA12/HLA-DQA1/ADH1B/BMP4/NDP/PLIN1/CX3CR1
cnetplot
cnetplot(y, foldChange=geneList)
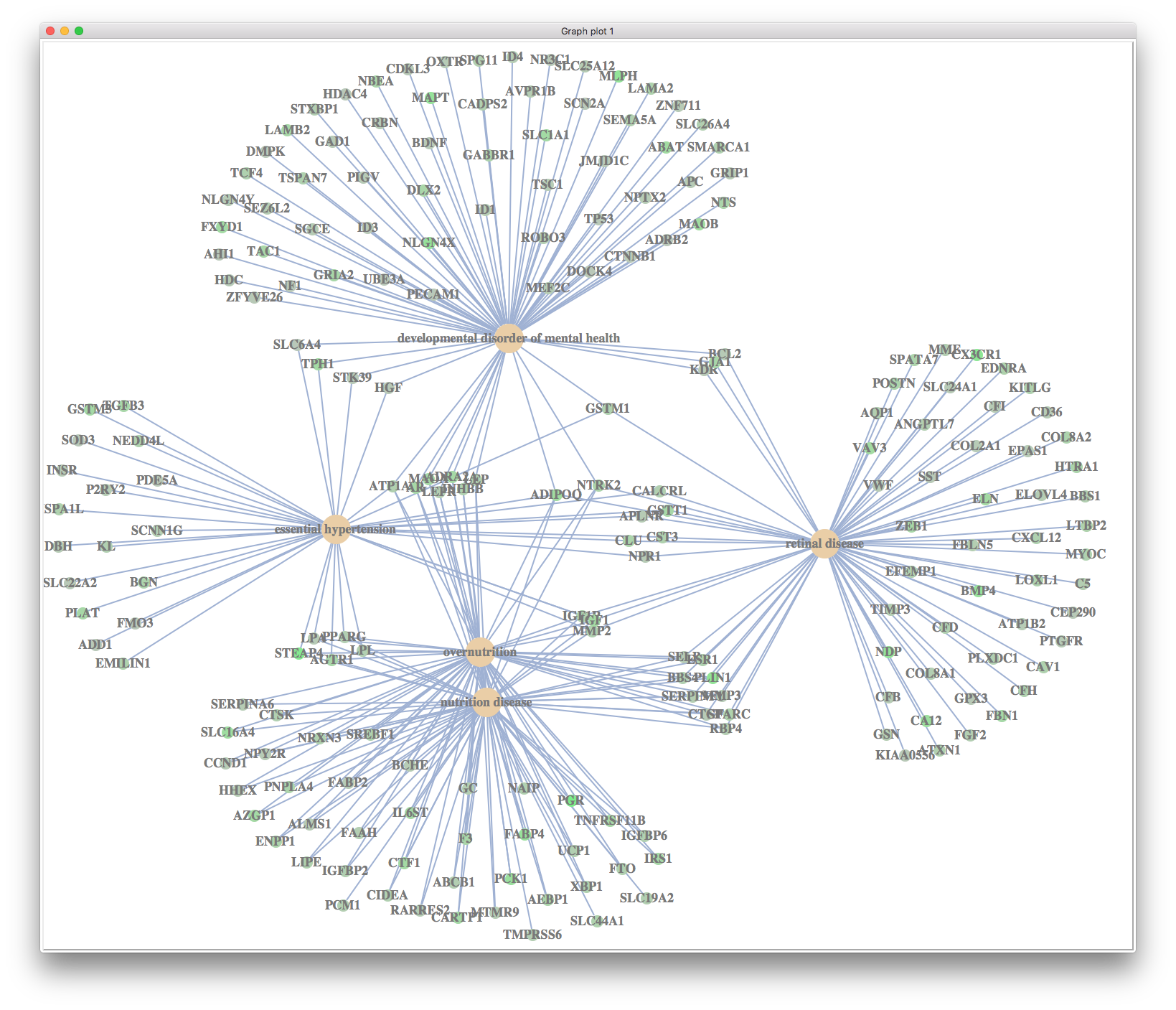
In the figure, we can find that most significant associated diseases are down-regulated.
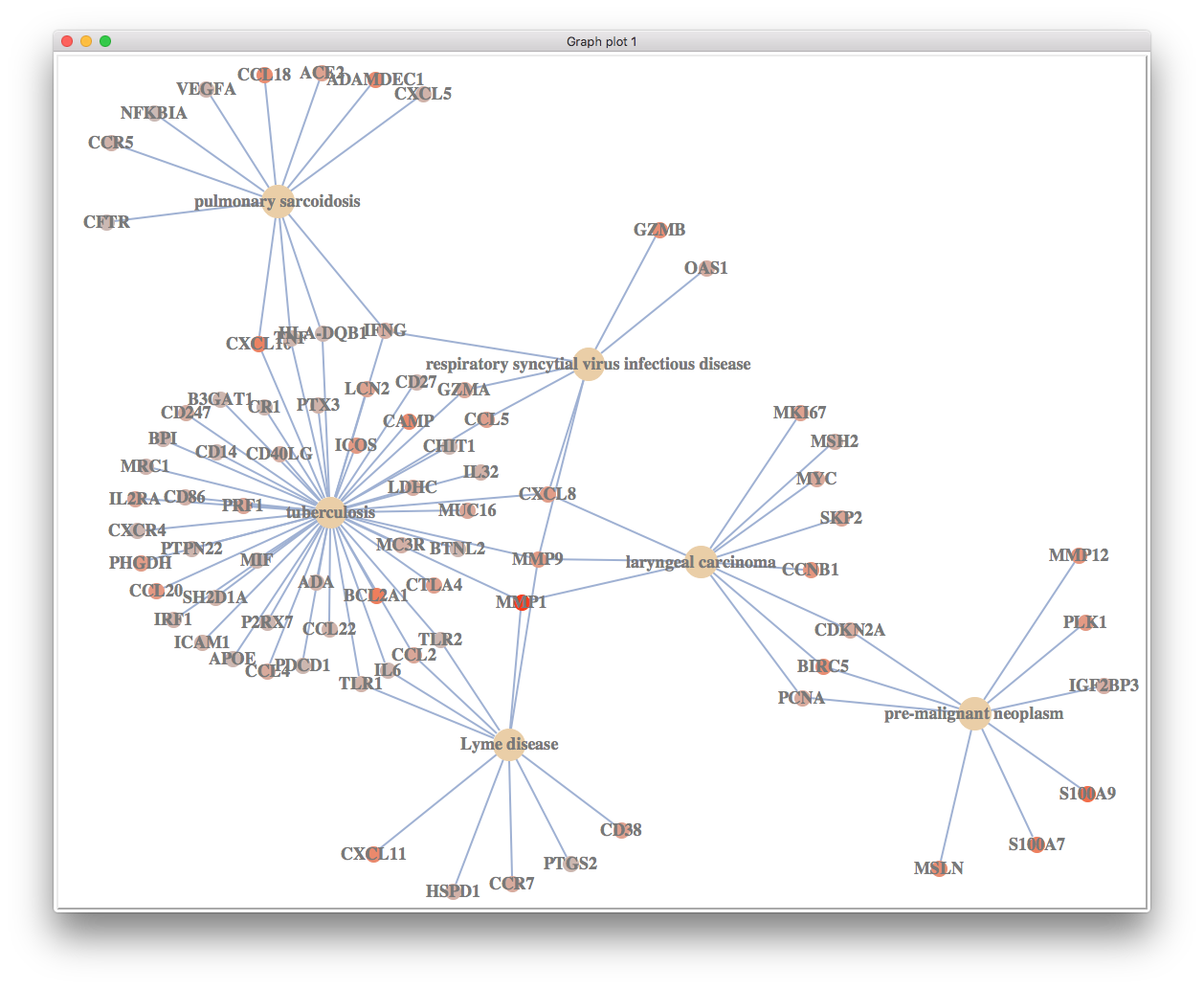
We can of course visualize significant up-regulated disease pathways.
up <- y$Description[order(y$NES, decreasing=TRUE)][1:6]
up
## [1] "tuberculosis"
## [2] "respiratory syncytial virus infectious disease"
## [3] "laryngeal carcinoma"
## [4] "Kawasaki disease"
## [5] "breast ductal carcinoma"
## [6] "pulmonary sarcoidosis"
cnetplot(y, foldChange=geneList, showCategory=up)
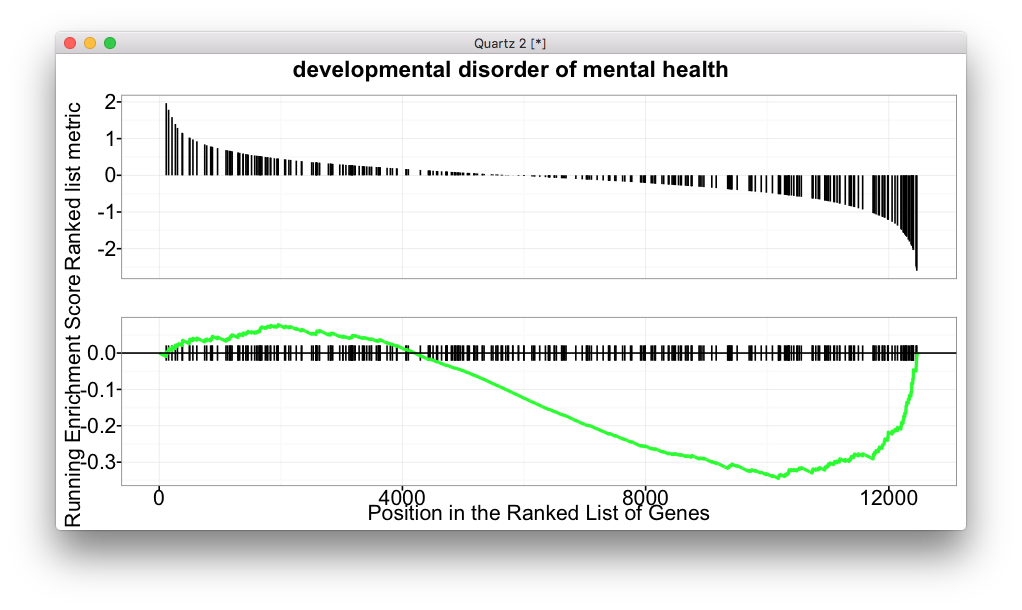
gseaplot
gseaplot now supports adding plot title.
gseaplot(y, y$ID[1], title=y$Description[1])
Citation
G Yu, LG Wang, GR Yan, QY He. DOSE: an R/Bioconductor package for Disease Ontology Semantic and Enrichment analysis. Bioinformatics 2015, 31(4):608-609.