rename phylogeny tip labels in treeio
require(treeio)## Loading required package: treeio## Registered S3 method overwritten by 'treeio':
## method from
## root.phylo ape## treeio v1.12.0 For help: https://yulab-smu.github.io/treedata-book/
##
## If you use treeio in published research, please cite:
##
## LG Wang, TTY Lam, S Xu, Z Dai, L Zhou, T Feng, P Guo, CW Dunn, BR Jones, T Bradley, H Zhu, Y Guan, Y Jiang, G Yu. treeio: an R package for phylogenetic tree input and output with richly annotated and associated data. Molecular Biology and Evolution 2020, 37(2):599-603. doi: 10.1093/molbev/msz240require(ggtree)## Loading required package: ggtree## ggtree v2.3.0 For help: https://yulab-smu.github.io/treedata-book/
##
## If you use ggtree in published research, please cite the most appropriate paper(s):
##
## [36m-[39m Guangchuang Yu. Using ggtree to visualize data on tree-like structures. Current Protocols in Bioinformatics, 2020, 69:e96. doi:10.1002/cpbi.96
## [36m-[39m Guangchuang Yu, Tommy Tsan-Yuk Lam, Huachen Zhu, Yi Guan. Two methods for mapping and visualizing associated data on phylogeny using ggtree. Molecular Biology and Evolution 2018, 35(12):3041-3043. doi:10.1093/molbev/msy194
## [36m-[39m Guangchuang Yu, David Smith, Huachen Zhu, Yi Guan, Tommy Tsan-Yuk Lam. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution 2017, 8(1):28-36. doi:10.1111/2041-210X.12628tr <- read.tree(text = "((a,(b,c)),d);")
genus <- c("Gorilla", "Pan", "Homo", "Pongo")
species <- c("gorilla", "spp.", "sapiens", "pygmaeus")
geo <- c("Africa", "Africa", "World", "Asia")
d <- data.frame(label = tr$tip.label, genus = genus,
species = species, geo = geo)
d## label genus species geo
## 1 a Gorilla gorilla Africa
## 2 b Pan spp. Africa
## 3 c Homo sapiens World
## 4 d Pongo pygmaeus Asiaggtree(tr) %<+% d + xlim(NA, 5) +
geom_tiplab(aes(label=paste0('italic(', genus, ')~bolditalic(', species, ')~', geo)), parse=T)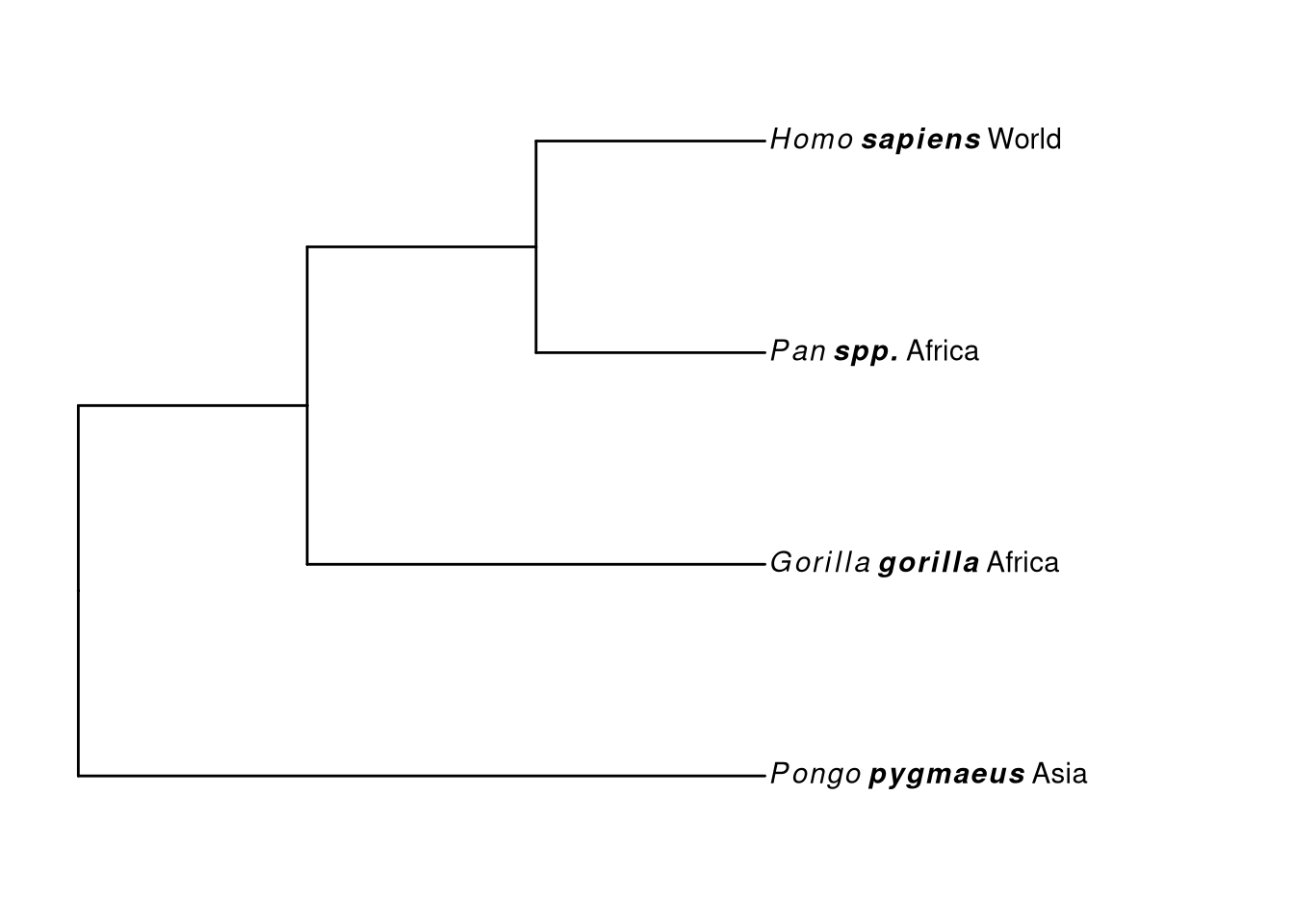
However, it is also possible to rename taxa of the tree object (either treedata or phylo) in treeio:
tr2 = rename_taxa(tr, d, label, genus)
write.tree(tr2)## [1] "((Gorilla,(Pan,Homo)),Pongo);"d2 = dplyr::mutate(d, newlab = paste(genus, species, sep='|'))
d2## label genus species geo newlab
## 1 a Gorilla gorilla Africa Gorilla|gorilla
## 2 b Pan spp. Africa Pan|spp.
## 3 c Homo sapiens World Homo|sapiens
## 4 d Pongo pygmaeus Asia Pongo|pygmaeustr3 = rename_taxa(tr, d2, label, newlab)
write.tree(tr3)## [1] "((Gorilla|gorilla,(Pan|spp.,Homo|sapiens)),Pongo|pygmaeus);"If the input tree object is a treedata instance, you can use write.beast to export the tree with associated data to a BEAST compatible NEXUS file.