msigdf + clusterProfiler全方位支持MSigDb
生信界的网红Stephen Turner在github上有个msigdf的包,我在他写这个包的时候,就写了个gist,连接clusterProfiler,我写gist的时候是2016年的8月,很高兴网红还惦记着我的gist。
msigdf这个包把著名的Broad Institute著名的Molecular Signatures Database (MSigDB)数据以data frame的形式打包成R包,这样子非常方便使用,当然他后来没有更新,而一个fork的版本,ToledoEM/msigdf把数据更新为最新版本v6.2,发布于2018年7月。
这个包,天生就方便我们clusterProfiler用户,我在《Comparison of clusterProfiler and GSEA-P》一文中,为了比较clusterProfiler和Broad Institute出品的GSEA-P软件,特意打包了一个gmt文件,这样方便在注释一样的情况下比较,有了这个示例,其实大家就应该知道,下载gmt文件,然后就可以用clusterProfiler分析了,也就是说clusterProfiler是支持MSigDb的,再一次敲重点,不要再以为clusterProfiler只做GO和KEGG了,clusterProfiler啥都能干。
好了,那么有这个包之后,我们连下载gmt文件都省了,直接载入这个包的数据,然后就可以无缝衔接clusterProfiler进行分析,还是原来的配方,还是熟悉的味道,你拥有了《enrichplot: 让你们对clusterProfiler系列包无法自拔》的各种可视化功能。
msigdf包
## devtools::install_github("ToledoEM/msigdf")
library(msigdf)
msigdf包含了人和鼠的数据,总共有以下8个类别:
| category | description |
|---|---|
| H | hallmark gene sets |
| C1 | positional gene sets |
| C2 | curated gene sets |
| C3 | motif gene sets |
| C4 | computational gene sets |
| C5 | GO gene sets |
| C6 | oncogenic signatures |
| C7 | immunologic signatures |
在这里我将过滤出C2拿来做分析:
library(dplyr)
c2 <- msigdf.human %>%
filter(category_code == "c2") %>% select(geneset, symbol) %>% as.data.frame
下面这就是我们拿到的C2的注释,一个data.frame:
> head(c2, 3)
geneset symbol
1 NAKAMURA_CANCER_MICROENVIRONMENT_UP COL1A2
2 NAKAMURA_CANCER_MICROENVIRONMENT_UP GPIHBP1
3 NAKAMURA_CANCER_MICROENVIRONMENT_UP RET
接下来是clusterProfiler的表演时间,我们还是使用DOSE包里的geneList数据来做演示,如果不知道怎么搞自己的geneList,请移步《听说你有RNAseq数据却不知道怎么跑GSEA》。
library(clusterProfiler)
data(geneList, package="DOSE")
这里涉及到一个问题,geneList里的基因是ENTREZID,而这里msigdf包里是SYMBOL,这个简单,无非是个基因ID转换的过程而已,一般而言,对于你自己的注释数据,你不应当有这个问题;假如有,也是一个转换步骤而已,这里做为演示,我把geneList的ID给转了,你当然也可以转注释c2里的ID。
> id = bitr(names(geneList), "ENTREZID", "SYMBOL", "org.Hs.eg.db")
'select()' returned 1:1 mapping between keys and columns
Warning message:
In bitr(names(geneList), "ENTREZID", "SYMBOL", "org.Hs.eg.db") :
0.41% of input gene IDs are fail to map...
> head(id, 3)
ENTREZID SYMBOL
1 4312 MMP1
2 8318 CDC45
3 10874 NMU
我们看到bitr的输出,有部分ID是没法转的,这些应该过滤掉,然后用新的ID去给geneList重命名即可,要注意ID要一一对应,不能搞错,我这里用了match,确保操作没问题:
> geneList = geneList[names(geneList) %in% id[,1]]
> names(geneList) = id[match(names(geneList), id[,1]), 2]
好了,现在轮到熟悉的clusterProfiler分析了,大家应该都不陌生,在下面的文章中都有介绍过:
用enricher做超几何分布检验:
de <- names(geneList)[1:100]
x <- enricher(de, TERM2GENE = c2)
用head瞄一眼结果:
> head(x, 3)
ID
ROSTY_CERVICAL_CANCER_PROLIFERATION_CLUSTER ROSTY_CERVICAL_CANCER_PROLIFERATION_CLUSTER
SOTIRIOU_BREAST_CANCER_GRADE_1_VS_3_UP SOTIRIOU_BREAST_CANCER_GRADE_1_VS_3_UP
SHEDDEN_LUNG_CANCER_POOR_SURVIVAL_A6 SHEDDEN_LUNG_CANCER_POOR_SURVIVAL_A6
Description
ROSTY_CERVICAL_CANCER_PROLIFERATION_CLUSTER ROSTY_CERVICAL_CANCER_PROLIFERATION_CLUSTER
SOTIRIOU_BREAST_CANCER_GRADE_1_VS_3_UP SOTIRIOU_BREAST_CANCER_GRADE_1_VS_3_UP
SHEDDEN_LUNG_CANCER_POOR_SURVIVAL_A6 SHEDDEN_LUNG_CANCER_POOR_SURVIVAL_A6
GeneRatio BgRatio pvalue
ROSTY_CERVICAL_CANCER_PROLIFERATION_CLUSTER 49/99 140/21142 5.021928e-83
SOTIRIOU_BREAST_CANCER_GRADE_1_VS_3_UP 46/99 151/21142 2.834141e-74
SHEDDEN_LUNG_CANCER_POOR_SURVIVAL_A6 58/99 456/21142 3.097882e-71
p.adjust qvalue
ROSTY_CERVICAL_CANCER_PROLIFERATION_CLUSTER 8.050151e-80 5.370820e-80
SOTIRIOU_BREAST_CANCER_GRADE_1_VS_3_UP 2.271564e-71 1.515520e-71
SHEDDEN_LUNG_CANCER_POOR_SURVIVAL_A6 1.655302e-68 1.104368e-68
geneID
ROSTY_CERVICAL_CANCER_PROLIFERATION_CLUSTER CDCA8/MCM10/CDC20/FOXM1/KIF23/CENPE/MYBL2/MELK/CCNB2/NDC80/TOP2A/NCAPH/E2F8/ASPM/RRM2/CEP55/DLGAP5/UBE2C/HJURP/APOBEC3B/PBK/NUSAP1/CDCA3/TPX2/TACC3/NEK2/CENPM/RAD51AP1/UBE2S/CCNA2/CDK1/ERCC6L/MAD2L1/GINS1/BIRC5/KIF11/EZH2/TTK/NCAPG/AURKB/CHAF1B/CHEK1/TRIP13/PRC1/KIFC1/KIF18B/KIF20A/DTL/AURKA
SOTIRIOU_BREAST_CANCER_GRADE_1_VS_3_UP CDC45/CDCA8/MCM10/CDC20/FOXM1/CENPE/MYBL2/MELK/CCNB2/NDC80/TOP2A/NCAPH/E2F8/ASPM/RRM2/CEP55/DLGAP5/UBE2C/HJURP/APOBEC3B/NUSAP1/CDCA3/TPX2/TACC3/NEK2/SLC7A5/CENPN/UBE2S/CCNA2/CDK1/MAD2L1/GINS1/BIRC5/KIF11/EZH2/TTK/NCAPG/AURKB/CHEK1/TRIP13/PRC1/KIFC1/KIF18B/QPRT/KIF20A/AURKA
SHEDDEN_LUNG_CANCER_POOR_SURVIVAL_A6 CDC45/NMU/CDCA8/MCM10/CDC20/FOXM1/KIF23/CENPE/MYBL2/MELK/CCNB2/NDC80/TOP2A/NCAPH/E2F8/ASPM/RRM2/CEP55/DLGAP5/UGT8/UBE2C/HJURP/APOBEC3B/SKA1/PBK/NUSAP1/CDCA3/TPX2/TACC3/NEK2/SLC7A5/CENPM/RAD51AP1/CENPN/UBE2S/CCNA2/CDK1/ERCC6L/MAD2L1/GINS1/KIF18A/CDT1/BIRC5/KIF11/EZH2/TTK/NCAPG/GPR19/AURKB/GINS2/CHEK1/TRIP13/PRC1/KIF18B/MMP12/KIF20A/DTL/AURKA
Count
ROSTY_CERVICAL_CANCER_PROLIFERATION_CLUSTER 49
SOTIRIOU_BREAST_CANCER_GRADE_1_VS_3_UP 46
SHEDDEN_LUNG_CANCER_POOR_SURVIVAL_A6 58
然后GSEA分析用GSEA函数:
y <- GSEA(geneList, TERM2GENE = c2)
同样我们也用head瞄一眼结果:
> head(y, 3)
ID
ONKEN_UVEAL_MELANOMA_DN ONKEN_UVEAL_MELANOMA_DN
NAKAMURA_TUMOR_ZONE_PERIPHERAL_VS_CENTRAL_DN NAKAMURA_TUMOR_ZONE_PERIPHERAL_VS_CENTRAL_DN
BONOME_OVARIAN_CANCER_SURVIVAL_SUBOPTIMAL_DEBULKING BONOME_OVARIAN_CANCER_SURVIVAL_SUBOPTIMAL_DEBULKING
Description
ONKEN_UVEAL_MELANOMA_DN ONKEN_UVEAL_MELANOMA_DN
NAKAMURA_TUMOR_ZONE_PERIPHERAL_VS_CENTRAL_DN NAKAMURA_TUMOR_ZONE_PERIPHERAL_VS_CENTRAL_DN
BONOME_OVARIAN_CANCER_SURVIVAL_SUBOPTIMAL_DEBULKING BONOME_OVARIAN_CANCER_SURVIVAL_SUBOPTIMAL_DEBULKING
setSize enrichmentScore
ONKEN_UVEAL_MELANOMA_DN 491 -0.3915581
NAKAMURA_TUMOR_ZONE_PERIPHERAL_VS_CENTRAL_DN 466 -0.3459209
BONOME_OVARIAN_CANCER_SURVIVAL_SUBOPTIMAL_DEBULKING 479 -0.3882366
NES pvalue
ONKEN_UVEAL_MELANOMA_DN -1.752063 0.001221001
NAKAMURA_TUMOR_ZONE_PERIPHERAL_VS_CENTRAL_DN -1.542803 0.001230012
BONOME_OVARIAN_CANCER_SURVIVAL_SUBOPTIMAL_DEBULKING -1.734615 0.001231527
p.adjust qvalues rank
ONKEN_UVEAL_MELANOMA_DN 0.01896267 0.01308232 3641
NAKAMURA_TUMOR_ZONE_PERIPHERAL_VS_CENTRAL_DN 0.01896267 0.01308232 2573
BONOME_OVARIAN_CANCER_SURVIVAL_SUBOPTIMAL_DEBULKING 0.01896267 0.01308232 2967
leading_edge
ONKEN_UVEAL_MELANOMA_DN tags=41%, list=29%, signal=30%
NAKAMURA_TUMOR_ZONE_PERIPHERAL_VS_CENTRAL_DN tags=33%, list=21%, signal=27%
BONOME_OVARIAN_CANCER_SURVIVAL_SUBOPTIMAL_DEBULKING tags=37%, list=24%, signal=29%
core_enrichment
ONKEN_UVEAL_MELANOMA_DN CTNNBIP1/DPY19L2P2/CNN3/ADCY6/TBX2/RPL11/KAT2B/C6orf48/SCAP/ETV1/HADHA/EIF3G/EIF1B/HFE/FAM184A/KDM6A/SPIN1/MBNL2/ABHD6/MID2/CSDE1/PRKAR1A/SLC35D2/BAG5/CSGALNACT1/TGOLN2/SOX9/NCKIPSD/ZNF185/QARS/DPYSL2/GABARAPL1/LRRC1/PCNP/SEC62/PEX6/TOP2B/ALDH9A1/FHL2/DLC1/RPL15/ENPP2/ZC3H13/USP4/ATXN7/ZSCAN12/EDNRB/SLC25A36/TWF1/ERBB3/MGEA5/OSBPL10/DSTN/SETD2/MFAP2/TMEM47/FAM86B1/NKTR/TFAP2A/TJP1/ADD3/NFE2L1/ECSIT/DAZAP2/MANSC1/RPL14/ACAA1/SNCA/SEMA6A/RPL29/CD200/TDRD3/APPL1/SDC4/ADRB2/GPR153/CTDSP2/VAMP3/GALC/RSL1D1/PREPL/PBX1/IP6K1/CTNNB1/GMPR2/PDHB/KANK2/DAB2/DBP/CPS1/SERPINB6/ENTPD1/GORASP1/PCLO/PTP4A2/PIK3R1/GLUL/RYBP/LETMD1/SCAMP1/PRCP/LAMA4/NEDD9/SPRY2/XPC/ASAH1/NEK4/GAS1/HBB/EIF4B/IMPDH2/PEG10/TSPYL1/CRBN/GOLGA4/LMCD1/MAGEH1/FSTL3/NRIP1/CCND1/HIST1H2AC/TOM1L1/NDN/GLCE/PALLD/BEX4/CREBL2/ITM2A/SPRY1/ZMAT3/CCDC28A/SOBP/KCNS3/SSBP2/ZBTB38/HSD17B8/ID4/ALDH6A1/IL11RA/C3orf14/CTSF/VWA5A/EFEMP2/RAB17/SNAP23/TIMP3/UBL3/SORBS1/DALRD3/SNRK/HSPA2/ZNF91/JAM3/ARMCX2/HNMT/GSTM2/SERPINI1/FAM129A/CADPS2/LPAR6/PDGFC/GSTM1/TXNIP/CCNG2/TRAK1/MTUS1/CBX7/SMARCA1/MPPED2/CAV1/CIRBP/NAP1L2/DYNC2H1/MZT2B/HEMK1/FYCO1/GPD1L/SLC7A8/PMP22/ZNF415/SLC39A6/POSTN/SYBU/GNG11/ZSCAN18/ZBTB20/LAMB2/LZTFL1/PLSCR4/CARTPT/GSTM3/AZGP1/PDGFD/ZNF423/RNASE4/KCNE4/PSD3/EMCN/PPP1R3C
NAKAMURA_TUMOR_ZONE_PERIPHERAL_VS_CENTRAL_DN ETS2/ADD3/SEC16A/ABCC3/TIA1/MACF1/RAB40B/DENND4C/C1RL/MXI1/SEMA6A/HBP1/WDR11/IGSF3/ZNF24/SERPINB1/GALC/RSL1D1/DPYD/FBXO9/NRN1/WSB1/ZNF638/ARNT2/AES/GATAD1/CPS1/NFATC2IP/MAVS/TRA2A/TTC31/BPTF/IGBP1/DTWD1/SRSF5/SLC2A10/LOXL2/STAT6/GLUL/RYBP/ITM2B/LETMD1/BNIP3L/EIF4A1/PHF3/COL6A1/CAST/WFDC2/BCL6/LMO2/IL1R1/AUH/INSR/TESC/GAS1/TAF9B/HP/TNFSF10/DOK1/PIGV/PCDH7/SGSM2/CLEC2B/DUSP1/FUCA1/USP34/HSD17B11/SLC24A3/SNCAIP/AHR/MKNK2/RNF125/FZD10/ZNF451/HIST1H2AC/FGFR3/KLHL28/C2orf68/CFI/NR4A2/ARID5B/GOLGA8A/ASPH/ZNF44/ZNF226/BAG1/ATG14/NELL2/UNC5B/HEY1/ENOSF1/ORAI3/RCOR3/SEZ6L2/NBR1/FNDC3A/SH3BGRL/LUM/FOS/CITED2/SLC46A3/TSPAN1/SPOCK1/CCNG1/DCAF10/RAB31/RAB27B/SPG11/ZNF91/IL33/UGCG/RGL2/CLK4/HNMT/SORL1/LPAR6/ST6GALNAC2/AHNAK2/TXNIP/CCNG2/CFH/LRIG1/ZNF580/CLK1/TPBG/MEGF6/MZF1/SLC7A2/ZNF395/ATP8B1/TTC28/CPE/CCNL2/KDM4B/GRAMD1C/C16orf45/CHST15/N4BP2L2/NISCH/IKBKB/NOTCH2NL/PILRB/PLAT/PODNL1/MAOA/CROT/COL4A5/MUC1/SLC1A1/SYT17/STC2/STEAP4/TMC5/CYBRD1
BONOME_OVARIAN_CANCER_SURVIVAL_SUBOPTIMAL_DEBULKING STOM/SDC2/STAT5B/ENPP2/FRMD4A/PTPRD/SCAPER/DNAJB4/OBSL1/AVPR2/PTPRS/TCF15/PEX3/ASPSCR1/NR2F1/SMARCA2/MORC3/TTC37/SIRT1/SEMA3F/TLE3/SRSF11/CCNH/NR2F2/SETD2/TMEM47/TNKS/CDK14/BCAT2/ADCY9/LRRC8E/CRTC3/AKAP10/RANBP3/GSTM5/PPP3CB/PDE4A/TM2D1/RECK/SPOP/PCGF3/RASA1/ECE1/BTF3/SPRED2/RAD17/KCNQ1/KCNAB1/ERLIN2/RB1/MRPS27/PPP3CA/EXOC7/METTL3/WBP4/HDAC4/EPB41L3/AP1G2/FEZ1/HOXD3/TMEM168/CCPG1/EID1/PIK3R1/CREBBP/AMH/MAN2A1/ITM2B/BNIP3L/SCAMP1/NID1/UBE2B/KDR/LIMCH1/ASAH1/NHLRC2/STK39/WNT5A/GPR39/FBXO38/FILIP1L/WFS1/ACSL3/PDS5B/LINC00260/HOOK2/ZNF235/F2R/PSD4/GULP1/COL15A1/SLC24A3/APC/SNCAIP/GATM/SFI1/CAMK2N1/NDN/PTPRN2/ARHGEF10/FBXL5/ITM2A/PDCD6/ZNF444/MEF2C/RPS6KA2/RARRES2/LPAR1/EDEM3/FNDC3A/DCHS1/ZFYVE16/LEPR/REEP5/THSD7A/RUNX1T1/ZNF839/COL21A1/UBL3/JAM2/SNAI2/GIPC2/PJA2/SPOCK1/CDH13/SOCS2/SYNE1/NBL1/MYOF/JAM3/PNISR/PDE1A/TRPC1/KL/ANKRA2/FAM172A/FAT4/PTGER3/EFEMP1/TSPAN7/IRAK3/SESN1/CAV1/MKL2/SEMA3C/EDNRA/ZBTB16/TGFB1I1/SPATA7/NBR2/SPDEF/CCDC106/ECM2/ABCG2/MEOX2/IKBKB/AGTR1/ZEB1/GPRASP1/DKK2/IGFBP4/SPARCL1/IRS1/AMIGO2/LMOD1/DCN/MAOA/IL6ST/ZCCHC24/PDZRN3/PDGFD/MAOB/RNASE4/C7/PSD3/CILP/ABCA8
用clusterProfiler分析有多种好处,一个是你可以用compareCluster比较不同的条件,还有就是有非常好的可视化函数,《enrichplot: 让你们对clusterProfiler系列包无法自拔》这篇文章你不容错过。
比如:
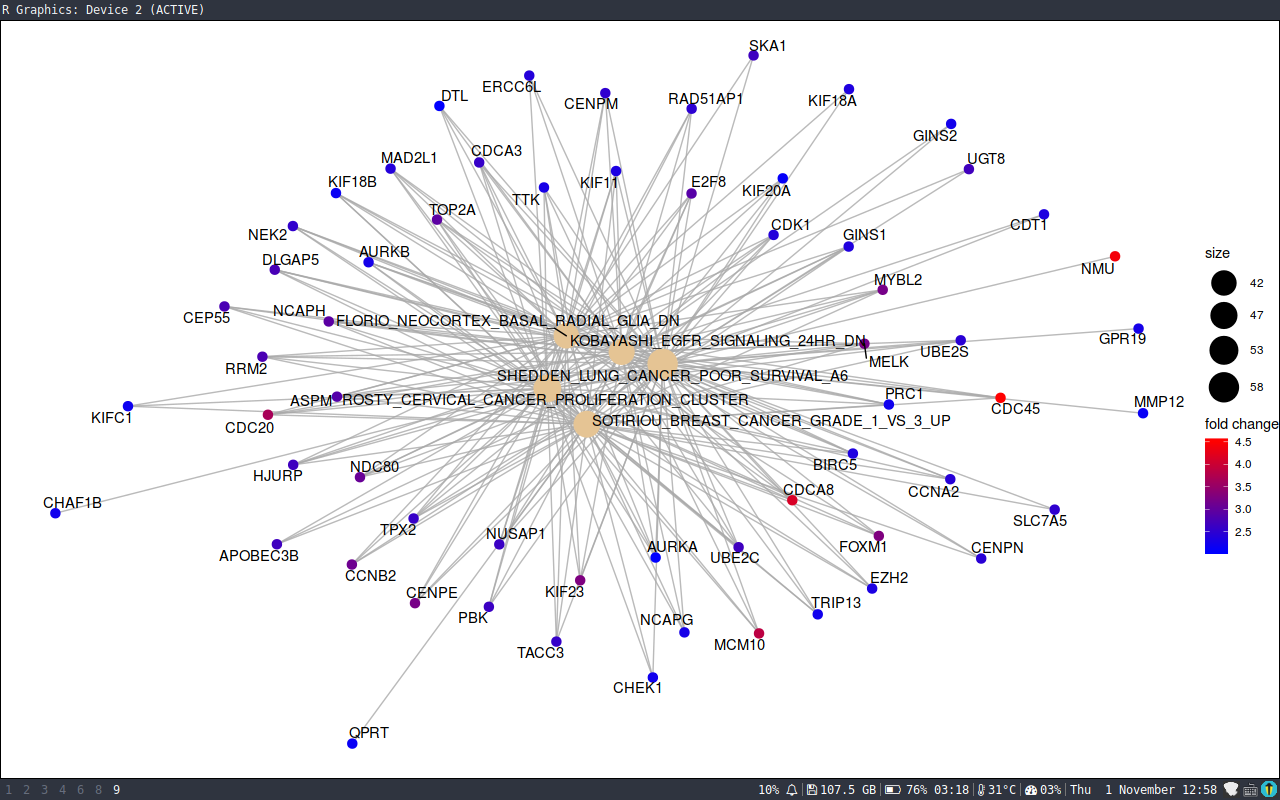
> cnetplot(x, foldChange=geneList)
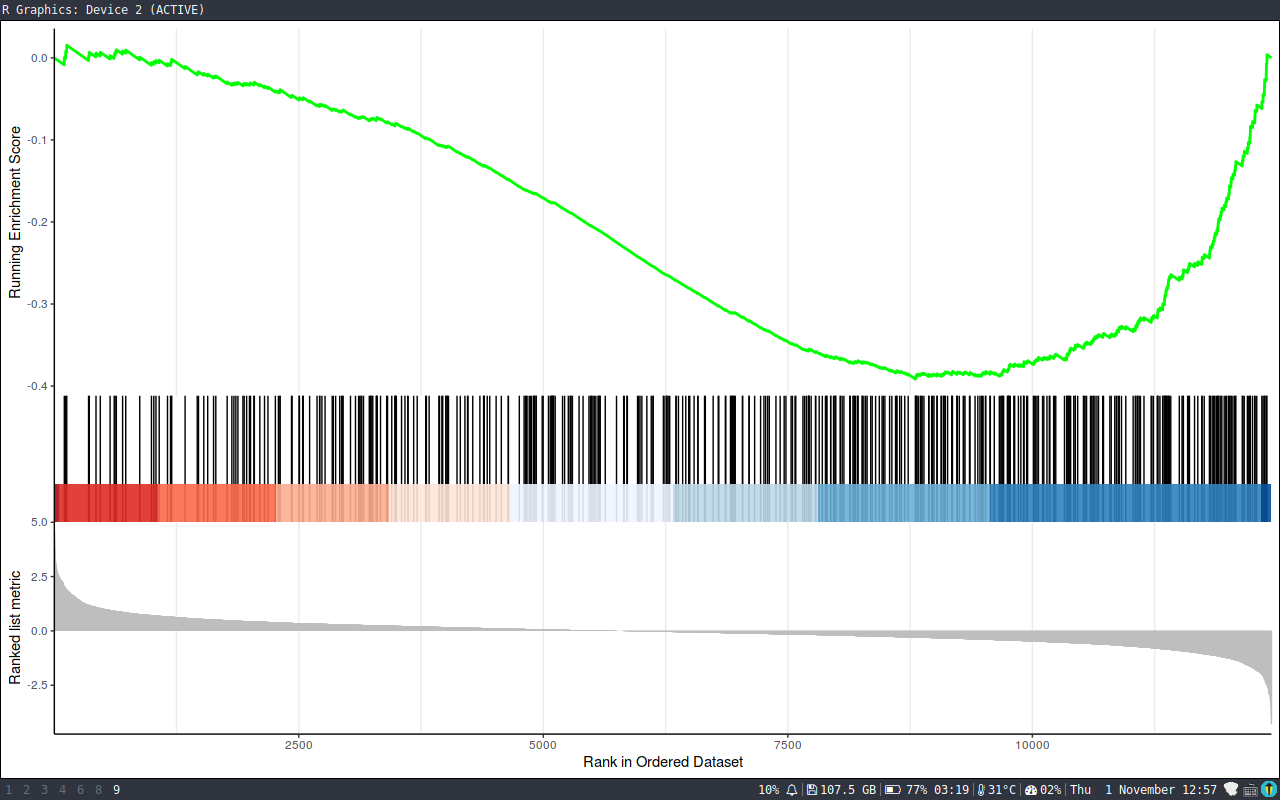
> gseaplot2(y, 1)