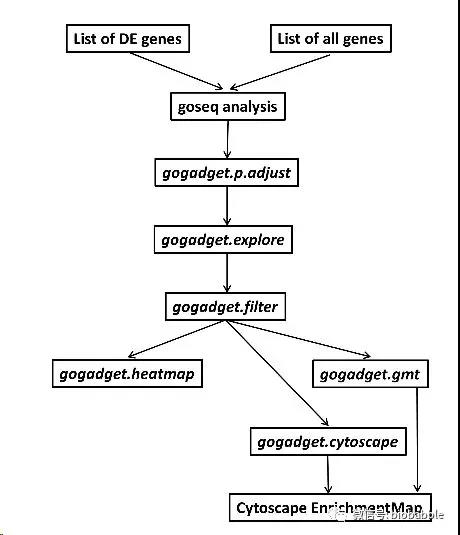
Hello! First of all, I would like to thank you for this wonderful and very powerful package!
I have tried to plot a phylogenetic tree with heatmap of associated matrix (with
gheatmap). I found that the row names of matrix doesn’t exactly match to the tip names of tree in case if there is missing data in associated matrix. In other words, if we have two species (e.g., Species198 and Species1981), but only one of them is represented in the associated matrix, we will have colored cells for both species in the heatmap.