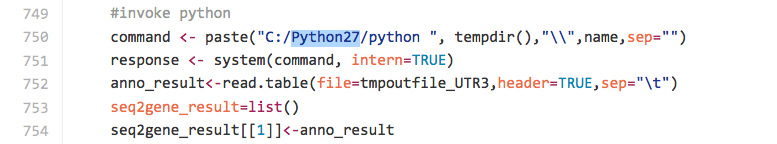
Subsetting is commonly used in ggtree as we would like to for example separating internal nodes from tips. We may also want to display annotation to specific node(s)/tip(s).
Some software may stored clade information (e.g. bootstrap value) as internal node labels. Indeed we want to manipulate such information and taxa labels separately.
In current ggplot2 (version=1.0.1, access date:2015-09-23), it support subset. For instance: