FigTree is designed for viewing
beast output as demonstrated by their example data:
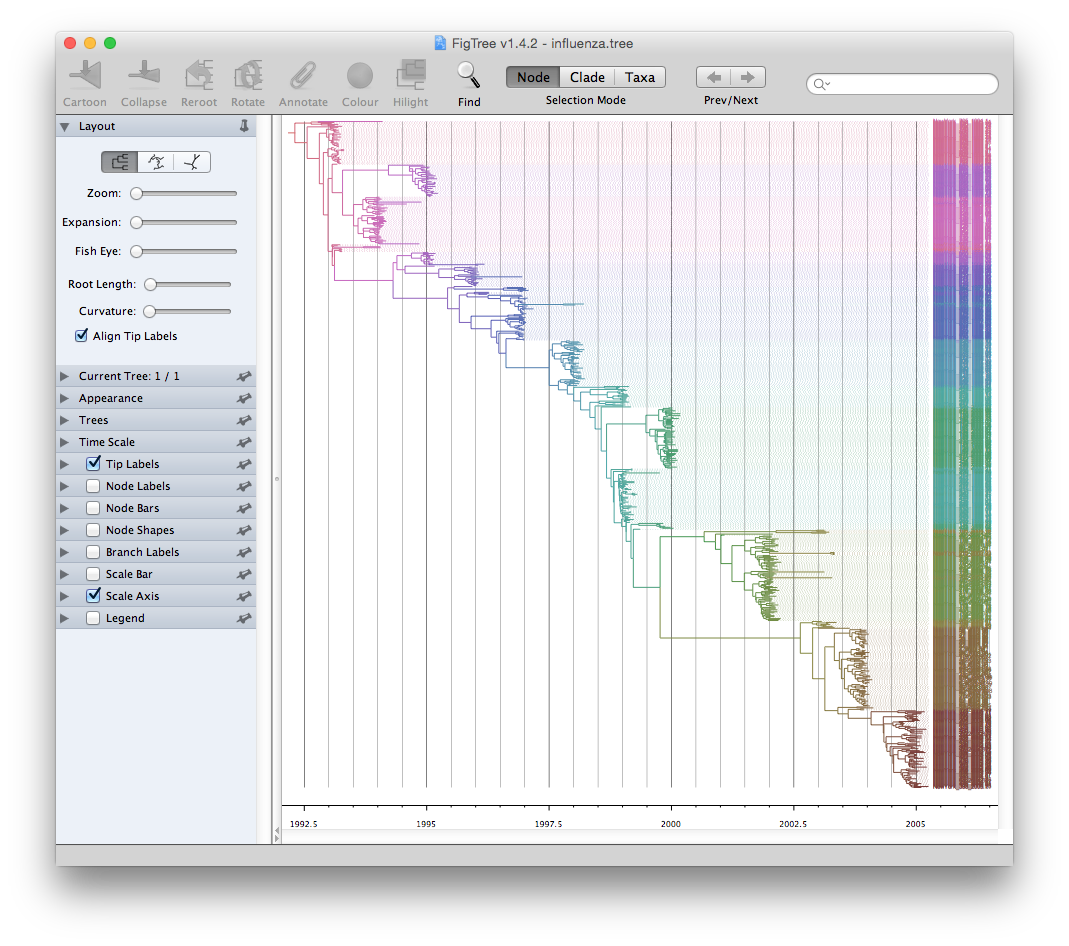
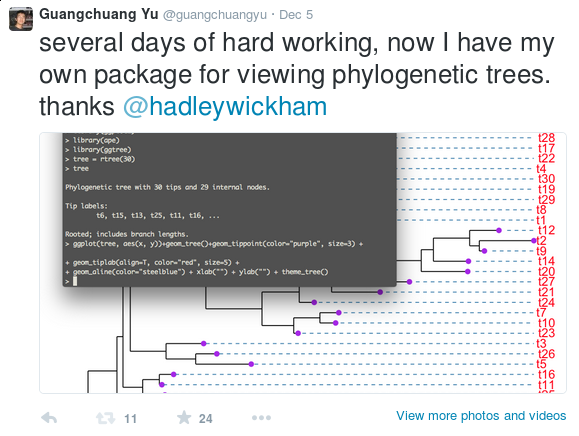
When I need to annotate nucleotide substitutions in the phylogenetic tree, I found that all the software are designed to display the tree but not annotating it. Some of them may support annotating the tree with specific data such as bootstrap values, but they are restricted to a few supported data types. It is hard/impossible to inject user specific data.
Genetic drift is the term used in population genetics to refer to the statistical drift over time of gene frequencies in a population due to random sampling effects in the formation of successive generations. In a narrower sense, genetic drift refers to the expected population dynamics of neutral alleles (those defined as having no positive or negative impact on reproductive fitness), which are predicted to eventually become fixed at zero or 100% frequency in the absence of other mechanisms affecting allele distributions.
The most important keyword in the definition of genetic drift is random sampling effects. The figure belowed illustrates this idea. The surviving individuals do not necessarily have selection advantage. They are randomly selected.