In ggtree, we provide a function annotation_image for annotating phylogenetic tree with images.

In ggtree, we provide a function annotation_image for annotating phylogenetic tree with images.
ChIPseeker is an R package for ChIP peak annotation, comparison and visualization.
We have implemented several visualization methods, including vennpie that was designed for viewing annotation overlap as shown below:
flip function is a feature request from ggtree user. It accepts a tree view and two node numbers and exchange the positions of the selected clades.
require(ggtree)
set.seed(2015-07-01)
tr <- rtree(30)
p <- ggtree(tr) + geom_text(aes(label=node))
gridExtra::grid.arrange(p, flip(p, 38, 33), ncol=2)
This is a feature request from clusterProfiler user. It’s similar to what I implemented in clusterProfiler for comparing biological themes. For comparing different enrichment results, the x-axis represent different gene clusters while for a single enrichment result, the x-axis can be gene count or gene ratio. This is actually similar to traditional barplot, with dot position as bar height and dot color as bar color. But dotplot can represent one more feature nicely by dot size and it can be a good alternative to barplot.
showtext is a neat solution to use various types of fonts in R graphs and make it easy to use funny fonts. With showtext, we can draw phylogenetic tree with different types of fonts even with symbolic/icon fonts.
require(showtext)
font.add.google("Gochi Hand", "gochi")
font.add.google("Rock Salt", "rock")
link = "http://img.dafont.com/dl/?f=wm_people_1";
download.file(link, "wmpeople1.zip", mode = "wb");
unzip("wmpeople1.zip");
font.add("wmpeople1", "wmpeople1.TTF");
link = "http://img.dafont.com/dl/?f=emoticons";
download.file(link, "emoticons.zip", mode = "wb");
unzip("emoticons.zip");
font.add("emoticons", "emoticons.ttf");
showtext.auto()
set.seed(2015-05-14)
library(ggplot2)
library(ggtree)
tree <- rtree(30)
ggtree(tree, color="darkgreen") + geom_tiplab(family="gochi") + theme_classic() +
theme(axis.text.x=element_text(size=rel(4), family="emoticons", color="firebrick")) +
scale_y_continuous(breaks=seq(0, 30, 4), labels=letters[c(12:17, 20, 21)]) +
theme(axis.text.y=element_text(size=rel(4), family="wmpeople1", color="#FDAC4F")) +
annotate("text", x=2, y=18, label="Have fun with ggtree!",
family="rock", angle=30, size=12, color="steelblue")
I found source code plagiarism a year ago and reported this case to BMC Systems Biology:
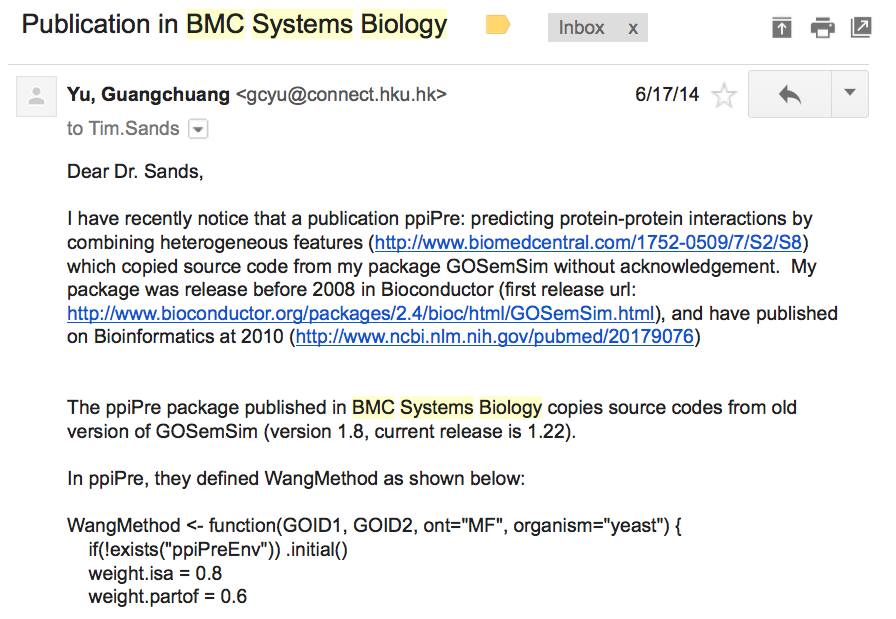
I listed source code of many functions that are exactly copied from GOSemSim with only function name changed in my email. The detail of source code plagiarism can also be found at Proper use of GOSemSim.