I have 8 packages published within the Bioconductor project.
A new package treeio was included in BioC 3.5 release.
I have 8 packages published within the Bioconductor project.
A new package treeio was included in BioC 3.5 release.
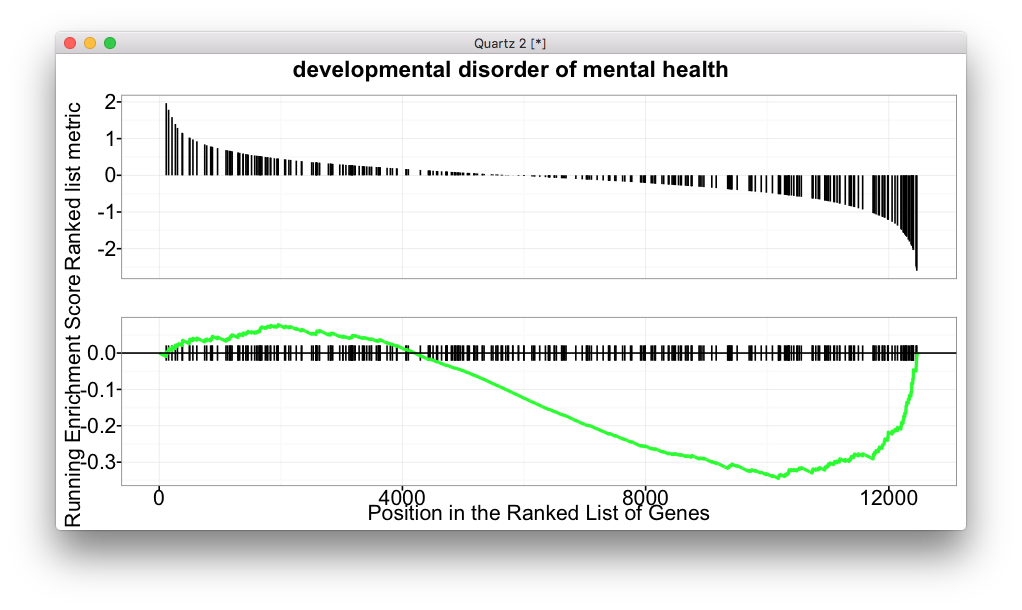
For GSEA analysis, we are familar with the above figure which shows the running enrichment score. But for most of the software, it lack of visualization method to summarize the whole enrichment result.
In DOSE (and related tools including clusterProfiler, ReactomePA and meshes), we provide enrichMap and cnetplot to summarize GSEA result.
I have 7 packages published within the Bioconductor project.
A new package meshes was included in BioC 3.4 release.
Leading edge analysis reports Tags to indicate the percentage of genes contributing to the enrichment score, List to indicate where in the list the enrichment score is attained and Signal for enrichment signal strength.
It would also be very interesting to get the core enriched genes that contribute to the enrichment.
Now DOSE, clusterProfiler and ReactomePA all support leading edge analysis and report core enriched genes.
发现Youtube上有一个视频叫Evolution of clusterProfiler, 是Landon Wilkins用Gource做的。于是我也来玩一下,看一下自己这几年码代码的过程。
Today is my birthday and it happened to be the release day of Bioconductor 3.3. It’s again the time to reflect what I’ve done in the past year.
In BioC 3.2 release, all my packages including GOSemSim, clusterProfiler, DOSE, ReactomePA, and ChIPseeker switch from Sweave to R Markdown for package vignettes.
To make it consistent between GOSemSim and clusterProfiler, ‘worm’ was deprecated and instead we should use ‘celegans’. As usual, information content data was updated.